Fig 2. Molecular representation of the PDB 4A7L complex, displaying an actin fiber (blue and green monomers, with ADP molecules in purple) with myosin heads (brown monomers).
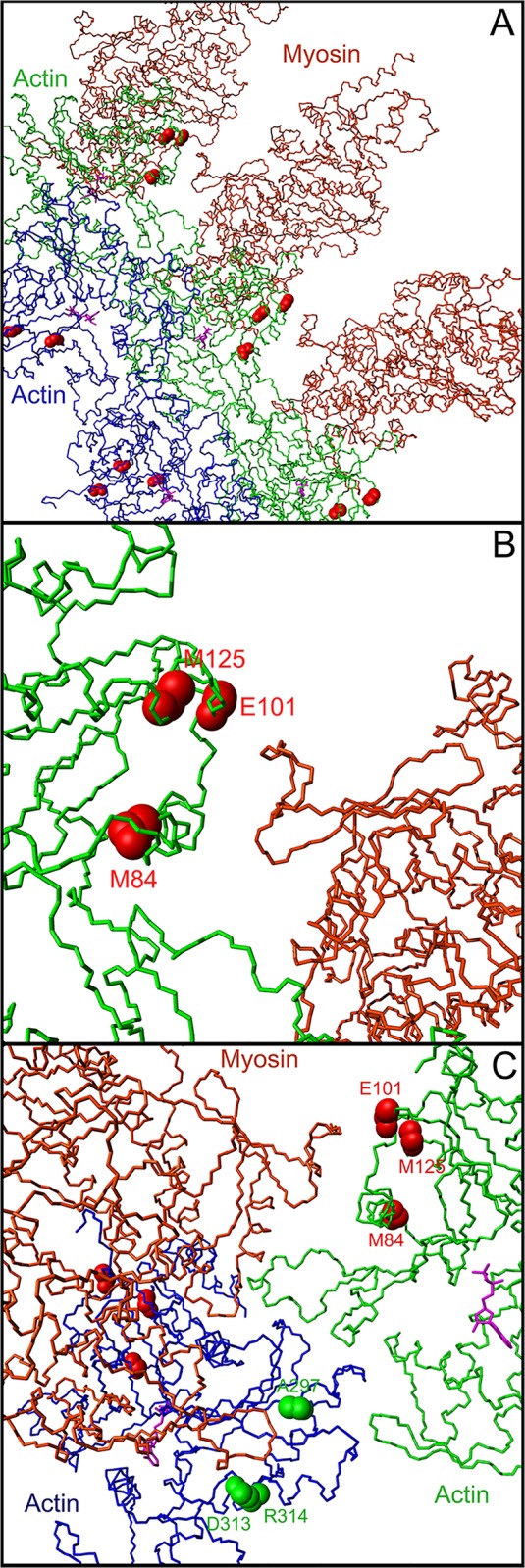
Actin and myosin amino acid numbers are according to human numbering (A) Backbone representation of an actin fiber in complex with 3 myosin heads, showing secondary structure elements. The backbone atoms of the 3 amino acids altered by mutations causing atrial septal defects (residues 84, 101, and 125) are shown by red spheres on every actin monomer. (B) Close up on the interaction between the region spanning residues 84, 101 and 125 of an F-actin monomer and a very close loop of the myosin head. The actin monomer is shown in green, the myosin head is shown in brown. The interatomic distances measured in the complex between residues 84, 101 and 125 and the myosin surface typically range from 3 to 10 Å. The 562–571 region of the myosin head makes numerous contacts with the surface of the actin filament, and interacts closely with residues 84, 101 and 125 on the surface of actin. (C) Close up showing one myosin head interacting with the region 84, 101, and 125 region of the actin monomer, and the 297, 313 and 314 region of an adjacent actin monomer. The 562–571 region of the myosin head closely interacts with residues whose mutation leads to atrial septal defects (84, 101, and 125, in red), whereas the 367–365 region (human numbering) of the same myosin head interacts directly with an adjacent actin monomer (residues 297, 313, 314, in green), whose mutation leads to cardiomyopathies. The orientation of the actin monomers in panel A and B is similar whereas the molecules in panel C have been rotated for a better view of the interaction with residues 297, 313, and 314.
