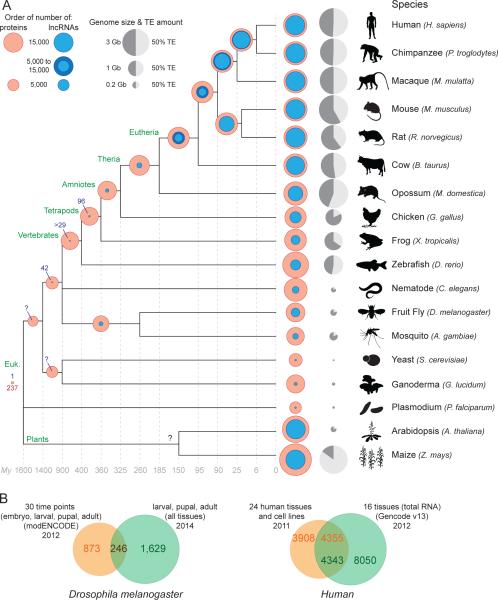
Figure 1. Rapid turnover of lncRNA repertoires.
A. Evolution of lncRNA and coding gene content. The amounts of lncRNA (blue circle; see below for references) and protein-coding coding (red circles) genes are superimposed to facilitate their comparison. Transposable element (TE) content and genome size are represented for each species (0% for Plasmodium [160]) as a grey circle next to the species name. The light gray fraction represents TE content, and the size of the circle reflects the size of the genome. The number of conserved orthologous genes is shown at each tree node when estimates are available or can be inferred from the literature (see below for references). Shared lncRNA amounts in tetrapods are from [3] and the pan-vertebrate lncRNA count (n=29) is from [12]. In eutherians (placental mammals), shared amounts are also extrapolated from [60, 63] and variations between studies are shown using a darker blue circle. The amount of shared lncRNA genes between Drosophila and mosquito is extrapolated from [67] and the 42 syntenic lncRNAs between Drosophila and vertebrates is from [5]. Beyond ribosomal RNA genes, we are only aware of a single lncRNA conserved across nearly all eukaryotes, the telomeric RNA TERRA [161-163].
References for lncRNA genes amounts are as follow: human, Gencode v19, Dec 2013, GRCh37 - Ensembl 74 [2] and [3, 164]; chimpanzee, macaque [3]; mouse, Gencode v2, Dec 2013, GRCm38 - Ensembl 74 [2] and [3, 164]; rat and cow lncRNA content was estimated to be similar to related organisms based on consistent amounts from single tissue analyses (liver for rat [63], skin [165] and muscle [166] for cow [see also 167]) and data for the organs of other mammals [3]; opossum [3]; chicken [3, 167]; frog [3]; zebrafish [12, 164, 167, 168]; nematode [167, 169]; Drosophila [5, 6]; in mosquito, 633 lncRNAs were identified with a very strict cut offs for identification. Therefore, given these first estimations for lncRNA content in drosophila, on the figure mosquito lncRNA content is represented as >1000 lncRNA genes (based on a set of 633 lncRNAs with very strict cut-offs [199]); yeast [167]; Ganoderma lucidum [170]; plasmodium [171]; Arabidopsis [7]; maize [8, 9]. Estimations from [3] include projected annotation, (see Extended Table 2 and Supp. Methods in ref. [3]). See also [4] for more details about most lncRNA datasets.
References for protein-coding genes amount for each species are from corresponding genome papers and updated using release 75 of Ensembl [172]. References for estimation of shared protein-coding genes are as follow: Eutherian [173-175]; Amniotes to Vertebrates [176-179], drosophila-Mosquito [180]; yeast to G. lucidum [181]; 237 P. falciparum proteins show strong matches to proteins in eukaryotic genomes [160]. B. Limited overlap between lncRNA catalogs obtained from different sources. The Venn diagrams show the amount of overlap in different lncRNA gene catalogs obtained for the same species. References: Drosophila melanogaster: [5, 6]. Human: [2, 27] [see 49].