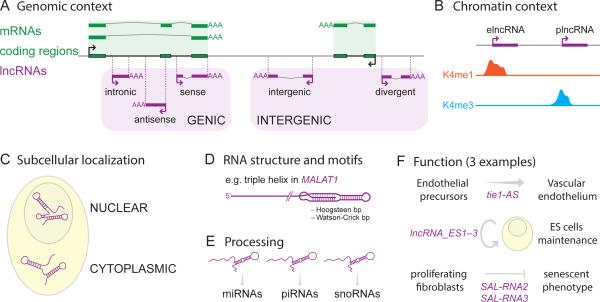
Figure 2. lncRNA classification.
LncRNA annotation is a challenging task under active development [reviewed in 182]. Here we illustrate a subset of many non-mutually exclusive criteria that may be used to classify lncRNAs. (A) Genomic context. lncRNAs may be divided based on their position and orientation relative to protein-coding genes: for instance overlapping (genic) or non-overlapping (intergenic: lincRNA) protein-coding genes [see 1, 11, 27].
(B) Chromatin context. Different populations can be defined by distinct chromatin marks around their transcription start site. For instance enhancer-associated (elncRNA) or promoter-associated (plncRNA) lncRNAs are characterized by mono- vs tri-methylation of lysine 4 of histone H3 respectively (K4me1 and K4me3) [29, 51]. This information can be combined with genomic context to further classify lncRNAs. For example, some intragenic lncRNAs, named meRNAs (multiexonic polyA+ RNAs), originate from active enhancers lying within protein-coding genes [110].
(C) Subcellular localization. Cellular fractionation and hybridization techniques can reveal whether lncRNAs are differentially located or accumulate in the nucleus or the cytoplasm [1] or other sub-organellar compartments such as nuclear paraspeckles [e.g. 183] or cytosolic ribosomal complexes [e.g. 26].
(D) RNA structure and motifs. Some lncRNAs may be grouped according to shared structural features and motifs. For instance, several lncRNAs, typified by MALAT1, are characterized by the formation of triple-helical structures at their 3’ end [184]. These structures and motifs are important for the stabilization, subcellular localization, and function of these lncRNAs. For example, a small motif involved in restricting lncRNA localization to the nucleus was identified [185].
(E) Processing. Some lncRNAs can be precursors of smaller RNA species such as piRNAs, miRNAs or snoRNAs [186-188]. For example, the BORDERLINE lncRNA is a precursor to small RNAs involved in demarcating an epigenetically distinct chromosomal domain in S. pombe [189]. It has also been shown that in yeast distinct lncRNA classes are sorted during 3’ end formation [190].
(F) Function. Reminiscent of Gene Ontology classification, lncRNAs may be grouped according to (i) their molecular activities (e.g. chromatin modification competitive endogenous loci [see 191 for review], architectural, etc.) or (ii) the cellular/biological processes they are involved in such as cell differentiation [e.g. 192], senescence [e.g. 193], circadian clock [e.g. 194], cell cycle regulation [reviewed in 195], pluripotency [e.g. 17, 31, 123], and innate immunity [196]. lncRNAs may also be classified based on their association with certain disease groups or states, such as neurological disorders [reviewed in 197] or cancer [198].