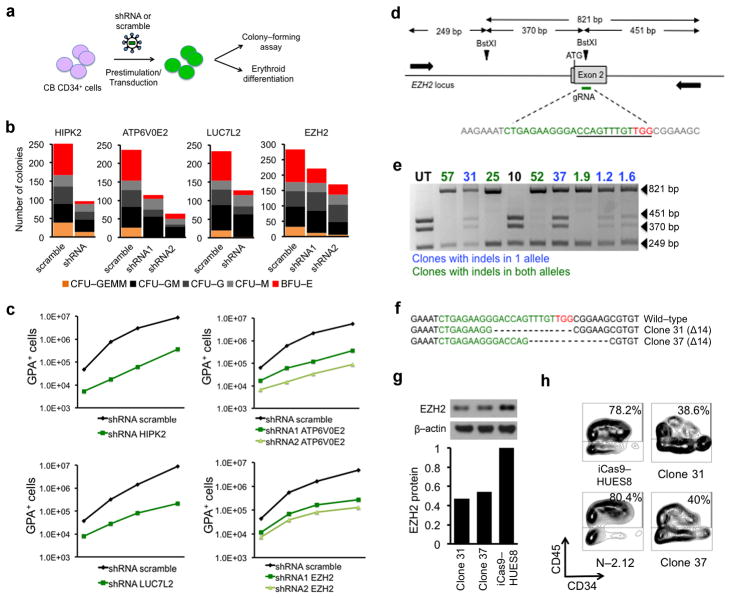
Figure 6. Validation of chr7q haploinsufficient genes.
(a) Experimental scheme. CB CD34+ cells were prestimulated for 48 h and transduced with a lentiviral vector encoding shRNA or scramble. 48 h later the cells were plated in colony-forming assays or in liquid erythroid induction culture.
(b) Methylcellulose assays of CB CD34+ cells expressing shRNAs against HIPK2, ATP6V0E2, LUC7L2, EZH2, or scramble, as indicated. Number of colonies from 1000 seeded cells is shown. The mean of duplicate experiments (from two independent transductions) is shown. (CFU-GEMM: colony-forming unit-granulocyte, erythrocyte, monocyte, megakaryocyte; CFU-GM: colony-forming unit-granulocyte, monocyte; CFU-G: colony-forming unit-granulocyte; CFU-M: colony-forming unit-monocyte; BFU-E: burst-forming unit-erythrocyte)
(c) Absolute number of GPA+ cells in vitro generated from CB CD34+ cells expressing shRNAs against HIPK2, ATP6V0E2, LUC7L2, EZH2 or scramble in erythroid media. Absolute numbers of GPA+ cells were calculated from the total cell counts and the percentage of GPA+ cells by flow cytometry. The mean values of duplicate experiments from two independent transductions are shown.
(d) Schematic representation of the EZH2 locus with the position of the gRNA target sequence, the BstXI restriction sites and the primers used for screening of CRISPR/Cas9-targeted alleles indicated. The gRNA sequence is shown in green and the PAM motif in red. The sequence of the BstXI restriction site is underlined. The beginning of the gRNA sequence is 23 nt downstream of the ATG.
(e) Representative image of RFLP analysis for screening of single cell clones. The 821 band (resistant to BstXI digest) indicates the presence of indels. Clones with indels in only one allele are shown in blue and clones with indels in both alleles are shown in green. Clones with both alleles intact are shown in black.
(f) Sequences of the wild-type EZH2 locus and the two clones (clone 37 and clone 31) harboring monoallelic indels, as indicated. Sequences shown were obtained from the 821 bp band and therefore correspond to the disrupted allele. The other allele was confirmed to be intact by sequencing the 370 and 451 bp bands (not shown). Each clone harbors a different 14-nucleotide deletion introducing a frame-shift.
(g) Western blot analysis (upper panels) and quantification (lower panels) of EZH2 expression in the two clones harboring monoallelic inactivation of EZH2, compared to that in the parental hESC line HUES8.
(h) Day 14 of hematopoietic differentiation of the two clones with monoallelic inactivation of EZH2, compared to that of the parental hESC line HUES8 and of normal iPSCs. One of two replicate experiments is shown.