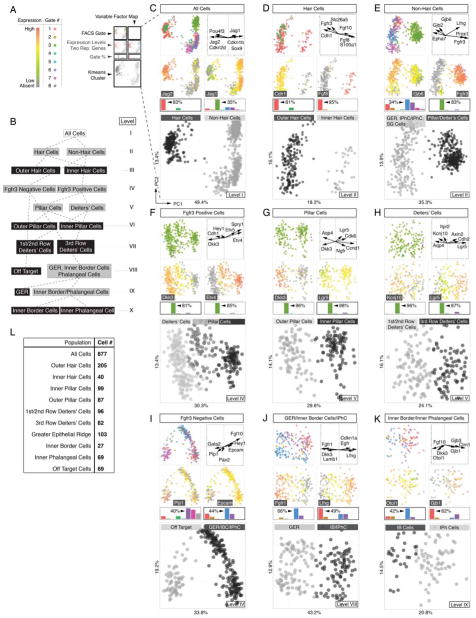
Figure 2.
Sequential clustering of organ of Corti cell types into cell type specific groups. (A) Legend and color key for panels C-K. (B) Subcluster tree of identified populations of all sorted cells across ten levels (I – X); grey boxes indicate intermediate groups and black boxes indicate final groups. (C) Principal component analysis (PCA) and k-means clustering of all 877 cells. Shown are projections onto lower-dimensional data space (principal component scores (PC1 and PC2)) visualizing different color-coded parameters. Each data point represents a single cell. Upper left: FACS-Gates 1 – 8. Upper right: Variable factor map indicates six representative genes that are most contributive for data separation. This representation shows the correlation between the first two components and the original variables (genes), which are called component loadings. Analogous to correlation coefficients their negative/positive values and direction can be understood as a measure of how much of variation in a variable is explained by the component. Second row: For each population expression levels of one representative marker is projected onto data, here Jag2 and Jag1. Gene expression levels range from grey (absent), green (low expression) to red (high expression). Bar graphs indicate k-means cluster-related relative contribution for each FACS gate (in %). Highest gate contribution for each group is indicated. Large square plot: Cells are colored according to computed k-means clusters (black = hair cells, grey = non-hair cells). (D-K) Analogous data representation for subsequently clustered subpopulations. (L) Table summarizing total cell number determined for each identified population. Related to Figures S2, S3 and S6.