Table 1.
Coronavirus spike (S) S1/S2 cleavage sites.
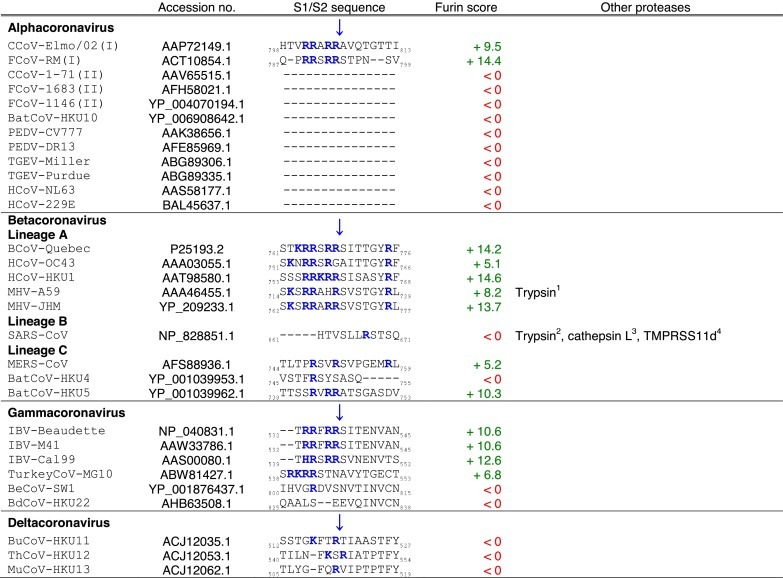
Tables. Analysis of coronavirus spike (S) cleavage sites.Table 1. Coronavirus S S1/S2 cleavage sites. The amino acid sequences of coronavirus S S1/S2 sites from the four coronavirus genera were aligned using ClustalX. Table 2. Coronavirus S S2′ cleavage sites. The amino acid sequences of coronavirus S S2′ sites from the four coronavirus genera were aligned using ClustalX. For Table 1, Table 2, the serotype of CCoV and FCoV are denoted in brackets (I or II). Table 3. Furin score and spike (S) amino acid sequence alignment of the S2′ region of passaged BCoV-B2.27.BO.P1 and other betacoronaviruses. Amino acid sequence alignment of S2′ region of S proteins of passaged BCoV-B2.27.BO.P1, BCoV-Quebec, MHV-JHM and SARS-CoV, based on sequence information by Borucki and collaborators (Borucki et al., 2013). Residues colored in red indicate positions with insertions/deletions. To generate furin scores in Table 1, Table 2, Table 3, sequences were queried into the PiTou 2.0 furin prediction algorithm that gives a score, with positive numbers (green) indicating predicted furin cleavage, while negative numbers (red) denote no predicted cleavage by furin. Furin scores that are borderline (<3) are denoted in grey. For comparison, the avian influenza strain A/muscovy duck/VietNam/209/2005(H5), which harbors the following polybasic cleavage site, R-R-R-K-R, has a +9.1 furin score. Other proteases, known to cleave coronavirus S1/S2 or S2′ sites are shown in the “Other proteases” column. Note that some proteases are known to cleave coronavirus S proteins, however, because the precise location of their cleavage site(s) has not been determined, they are not shown in Table 1, Table 2. Blue arrows denote the position of potential sites of cleavage. Basic arginine (R) and lysine (K) residues are highlighted in blue and bold font.
