Abstract
Purpose
Clinical testing for germline variation in multiple cancer susceptibility genes is available using massively parallel sequencing. Limited information is available for pre-test genetic counseling regarding the spectrum of mutations and variants of uncertain significance (VUSs) in defined patient populations.
Methods
We performed massively parallel sequencing using targeted capture of 22 cancer susceptibility genes in 278 BRCA1/2 negative patients with early onset breast cancer (diagnosed under age 40).
Results
Thirty-one patients (11%) were found to have at least one deleterious or likely deleterious variant. Seven patients (2.5% overall) were found to have deleterious or likely deleterious variants in genes for which clinical guidelines exist for management, namely TP53 (4), CDKN2A (1) MSH2 (1), and MUTYH (double heterozygote). Twenty-four patients (8.6%) had deleterious or likely deleterious variants in a cancer susceptibility gene for which clinical guidelines are lacking, such as CHEK2 and ATM. Fifty-four patients (19%) had at least one VUS, and six patients were heterozygous for a variant in MUTYH.
Conclusion
These data demonstrate that massively parallel sequencing identifies reportable variants in known cancer susceptibility genes in over 30% of patients with early onset breast cancer. However, only rare patients (2.5%) have definitively actionable mutations given current clinical management guidelines.
Keywords: early-onset breast cancer, cancer susceptibility, multiplex panel testing, massively parallel sequencing, genetic testing
INTRODUCTION
Mutations in the breast and ovarian cancer susceptibility genes, BRCA1 and BRCA2, are found in 10–20% of women with early-onset breast cancer (defined as breast cancer diagnosed under age 40)1. In comparison to women with postmenopausal breast cancer, women with early-onset breast cancer have a worse prognosis with increased recurrence rates, rates of distant metastases, and higher overall mortality1. As BRCA1/2 genetic testing is recommended for all women diagnosed with breast cancer under 402, further expansion of genetic testing to other moderate and high penetrance genes is commonly considered for this group. Further, it has the potential to identify women who may benefit from targeted breast cancer screening and prevention strategies aimed at decreasing morbidity and mortality, as has been demonstrated for BRCA1/2 mutation carriers3.
Beyond BRCA1/2, rare highly penetrant mutations in the genes CDH1, PTEN, STK11, and TP53 lead to cancer susceptibility syndromes4, for which the National Cancer Care Network (NCCN) provides guidelines for genetic testing and clinical management2. In addition to these high risk genes, breast cancer susceptibility is associated with rare moderate penetrance mutations in an increasing number of genes, such as ATM, CHEK2, and PALB2, which confer an approximately two to five fold increased risk of breast cancer4. Guidelines do not delineate patient populations for whom testing for mutations in moderate risk genes is expected to be beneficial, nor how the information of this testing should be applied in clinical management of cancer risk.
Despite these limitations, clinical testing based on massively parallel sequencing (MPS) is now commercially available for many known or proposed cancer susceptibility genes5,6. Rather than focusing on sequential testing of individual, well-studied genes due to defined clinical characteristics of the patient’s personal and family histories, these tests concurrently screen a large number of genes. A lack of data about the cancer risk and penetrance in women carrying these mutations has made the translation of potential life-saving strategies used in BRCA1/2 carriers to these women problematic7,8. Whereas frequencies of BRCA1/2 mutations are well studied, data is needed on the spectrum of variants in the other cancer susceptibility genes in defined patient populations. We report, herein, data on the frequency and type of variants in a panel of cancer susceptibility genes in BRCA1/2 negative women with early-onset breast cancer.
MATERIALS AND METHODS
Patient ascertainment
The study population was ascertained from academic and community hospital sites within the Penn Cancer Network and The Karmanos Cancer Institute at Wayne State University9. The majority of the patient population (253 patients, 91%) was ascertained via the Penn Cancer Network sixteen patients (6%) were from the Karmanos Cancer Institute at Wayne State University, and ascertainment data was not available for nine patients (3%). Acquisition of the patient samples was approved by the Institutional Review Boards of the corresponding institutions, and informed consent was obtained from each participant for use of their samples in genetic studies. Eligibility criteria for the study were: 1) diagnosis of breast cancer under age 40; 2) negative BRCA1/2 sequencing in a CLIA-approved laboratory; and 3) negative personal or family history of ovarian cancer. Analysis for BRCA1/2 large genomic rearrangements was not required, although negative clinical testing was available for 28% of patients.
DNA library preparation and sequencing
For each patient, one microgram of constitutional DNA was blunt ended and ligated with adaptors-embedded indexes. DNA quality, fragment size and concentration were measured with an Agilent 2100 Bioanalyzer. DNA libraries of sufficient quality were pooled pre-capture to 24-plex and hybridized to a custom designed Agilent SureSelect target library covering all coding exons and the flanking 10 base pairs of 22 genes. The genes included 20 study genes plus BRCA1 and BRCA2 and were: 1) high penetrance breast cancer susceptibility genes (CDH1, PTEN, STK11, TP53); 2) genes known to cause other cancer susceptibility (CDKN2A, MLH1, MSH2, MSH6, PMS2); 3) genes known or postulated to be moderate penetrance cancer susceptibility genes (ATM, BARD1, BRIP1, CHEK2, FAM175A,MRE11A, NBN, RAD50, PALB2, RAD51C); and 4) MUTYH, which leads to autosomal recessive polyposis.
Massively parallel sequencing data analysis
Raw sequencing data were aligned to the hg19 assembly of the human genome using Burrows-Wheeler Aligner (BWA) for short-read alignment (http://sourceforge.net/projects/bio-bwa/files/)10. BAM files were processed with Genome Analysis Toolkit (GATK) for detection of single nucleotide variants (SNVs) and insertion/deletion variants (indels) (http://www.broadinstitute.org/gatk/download)10,11 and annotated with ANNOVAR (http://www.openbioinformatics.org/annovar/annovar_download.html)10. Data was additionally analyzed using Pindel to improve sensitivity for medium sized indels (http://gmt.genome.wustl.edu/pindel/0.2.4/install.html)10 and xhmm for large genomic rearrangements (https://atgu.mgh.harvard.edu/xhmm/download.shtml)12. Quality control measures were calculated using Picard Tools (http://sourceforge.net/projects/picard/files/). Samples were sequenced to a mean coverage of 224×. Three samples were removed from the analysis for having >10% of targets with 0% coverage or <50% of targets with >10× coverage.
To identify all single nucleotide variants, small and medium sized insertion/deletions (indels) and large genomic rearrangements, variants were filtered to remove synonymous missense variants and intronic variants. Variants were removed from analysis if the alternate allele frequency was less than 0.2 and the total number of reads less than 20. All other insertion, deletions, nonsense variants, and splicing variants were retained for further analysis. Variants were kept for further analysis if found at an allele frequency of less than 0.1% in both the ESP6500 (http://evs.gs.washington.edu/EVS/) and 1000G (http://www.1000genomes.org/data) databases. Variants were analyzed if found at 0.1–1% allele frequency and previously reported to be a breast cancer susceptibility variant. Splicing variants were analyzed with Skippy and PupaSuite10,13. All variants were visually inspected in the Integrative Genomics Viewer (IGV, http://www.broadinstitute.org/software/igv/log-in)10.
Variant classification
In order to classify variants into a five-tiered system, a pipeline was developed which integrated posterior probability of pathogenicity data (when available), publically available database calls, protein position of the variant in a functional domain, in silico analysis10 of effect of variant on conservation with GERP, Siphy and PhyloP and functionality with SIFT, Polyphen2, LRT, MutationTaster and MutationAssessor (Supplementary Table 1). Specifically, variants were first assigned as a Variant of Uncertain Significance (VUS) if a) the posterior probability of pathogenicity > 0.0518 as recorded in the gene’s locus specific database (LSDB) if available or b) if the variant was not found in EVS6500, 1000 Genomes and dbSNP databases, if a LSDB was not available. If these conditions were not met, the variant was assigned as a likely benign Variant (i.e. if a) the posterior probability of pathogenicity < 0.0518 as recorded in the LSDB if available or b) if the variant was found in EVS6500, 1000 genomes or dbSNP databases, if a LSDB was not available). Exceptions were made for known pathogenic variants found in EVS6500, 1000G and dbSNP (i.e. CHEK2 c.1100delC). For the VUSs, variants were upgraded to deleterious variant if called pathogenic by two or more databases (HGMD http://www.hgmd.org/, Clinvar https://www.ncbi.nlm.nih.gov/clinvar/, and the LSDB of the gene (http://www.hgvs.org/dblist/glsdb.html). VUSs were upgraded to likely deleterious variants if at least four of the following five features (“D points”) indicated pathogenicity of the variant: 1) position of variant in a biologically important functional domain of the protein known to harbor pathogenic mutations; 2) pathogenic call in one database (HGMD, Clinvar, and the LSDB of the gene); 3) a normalized conservation score (NCS) of >2 (maximum 3); 4) a normalized functional score (NFS) of >4 (maximum 5); and 5) reported non-functional in a published in vitro assay. The Normalized Conservation Score was calculated by NCS = (GERPScore/x) + (PhyloPScore/x) + (SiPhyScore/x), where x=maximum score for each caller in the dataset. The Normalized Functional Score was calculated by NFS =(1-SIFTScore) + PP2HDIVScore + (1-LRTScore) + MutTasterScore + (MutAssessorScore/x), where x=maximum score for each caller in the dataset. If the NFS was between 3–4, the variant was given one D point if the AlignGVGD score (http://agvgd.iarc.fr/agvgd_input.php)14 was C55 or C65 or if the CONDEL score (http://agvgd.iarc.fr/agvgd_input.php)15 was “D”. For the likely benign variants, these variants were upgraded to VUSs if at least two features (“D points”, listed above) indicated pathogenicity of the variant. Likely benign variants were downgraded to benign variants if called a SNP by more than two databases (HGMD, Clinvar, dbSNP and the LSDB of the gene).
Validation of pipeline
In order to determine the efficiency and accuracy of our sequencing platform and bioinformatics and variant classification pipeline, we analyzed samples with variants identified by clinical sequencing in BRCA1, BRCA2, MSH2, or PALB2; these included two nonsense mutations, four indels, two large genomic rearrangements, and 34 single nucleotide variants. 100% of the 42 known variants were identified and correctly classified. For each identified deleterious and likely deleterious variant in a study sample, a separate stock aliquot of the patient's DNA sample from the aliquot used for MPS was used for Sanger sequencing of the genomic region containing the variant. Primers were developed using NCBI Primer Design software and PCR products were generated with Platinum Taq polymerase.
Statistical analysis of clinicopathogical variables
Statistical comparisons were made regarding the frequency of patients with certain clinical or pathological features within groups of patients as determined by variant status using a two-tailed Fisher’s exact test. Statistical comparisons of age, Penn II scores, and BOADICEA scores between groups of patients depending on variant status was performed using a two-tailed, type 2, Student’s t-test. Comparisons were run for deleterious/likely deleterious variant positive versus deleterious/likely deleterious variant negative (including the VUS positive patients in the latter group) and deleterious/likely deleterious variant positive versus deleterious/likely deleterious variant and VUS negative (excluding the VUS positive patients from both groups).
RESULTS
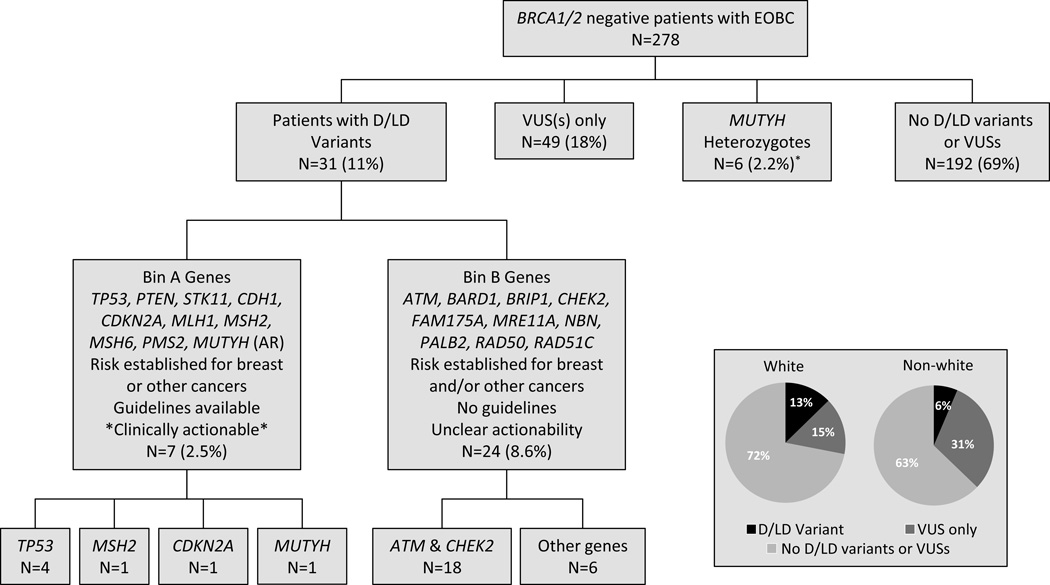
Characteristics of the early-onset breast cancer study population studied are shown in Table 1. Of the 278 patients, 169 (61%) had at least one variant found at <0.1% allele frequency in control public databases. After variant classification, 86 patients (31%) were found to have at least one deleterious variant, likely deleterious variant, or VUS (Figure 1). Thirty-one patients (11%) overall were identified to carry a total of 34 deleterious or likely deleterious variants, 53 patients (19%) had 57 VUSs (including 49 with a VUS only), and six patients (2.2%) were heterozygous for MUTYH variants.
Table 1.
Clinical and pathological characteristics of patients
| Characteristic | Study population (n=278) |
Mutation positivea (n=31) |
VUS Positiveb (n=55) |
Mutation/ VUS negativec (n=192) |
Mutation positive vs rest of populationd |
|---|---|---|---|---|---|
| Clinical characteristics | p-value | ||||
| Average age of onset of BC | 34 (20–39) | 34 (23–39) | 34 (24–39) | 34 (20–39) | NS |
| Self-reported race/ethnicity | NS | ||||
| White/Caucasian | 190 (69%) | 24 (77%) | 29 (53%) | 136 (71%) | NS |
| African American/Black | 66 (24%) | 5 (16%) | 19 (35%) | 42 (22%) | NS |
| Othere | 12 (4%) | 0 | 5 (9%) | 7 (4%) | NS |
| Not reported | 10 (4%) | 2 (7%) | 2 (4%) | 7 (4%) | NS |
| Ashkenazi Jewish | 27 (10%) | 6 (19%) | 3 (5%) | 18 (9%) | NS |
| Non-Jewish | 234 (84%) | 23 (74%) | 51 (93%) | 160 (83%) | NS |
| Personal cancer history | |||||
| Contralateral Breast Cancer | 36 (13%) | 6 (19%) | 5 (9%) | 25 (13%) | NS |
| 2nd primary malignancyf | 47 (17%) | 6 (19%) | 3 (5%) | 12 (6%) | 0.02 |
| Family cancer history | |||||
| Breast cancer | 188 (68%) | 25 (81%) | 35 (64%) | 129 (67%) | NS |
| Breast cancer age<40 | 76 (27%) | 9 (29%) | 15 (27%) | 52 (27%) | NS |
| Bilineal breast cancer | 34 (12%) | 7 (23%) | 5 (9%) | 22 (11%) | 0.08 |
| BRCA1/2 Prediction Models | |||||
| Penn II prior probability | 21% | 27% | 20% | 19% | 0.04 |
| BOADICEA | 15% | 29% | 13% | 14% | 0.005 |
| Pathological data | N (%) | N (%) | N (%) | N (%) | p-value |
| Ductal carcinoma in situ | 23/278 (8%) | 4/31 (13%) | 7/55 (13%) | 12/192 (6%) | NS |
| ER+ invasive BC | 147/214 (69%) | 19/22 (86%) | 29/44 (66%) | 99/148 (67%) | 0.09 |
| Her2+ invasive BC | 49/175 (28%) | 7/20 (35%) | 10/30 (33%) | 32/125 (26%) | NS |
| Stage I | 67/208 (32%) | 6/21 (29%) | 14/41 (34%) | 47/146 (32%) | NS |
| Stage IIA/B | 96/208 (46%) | 9/21 (43%) | 16/41 (39%) | 71/146 (49%) | NS |
| Stage IIIA/B/C | 43/208 (21%) | 6/21 (29%) | 11/41 (27%) | 26/146 (18%) | NS |
| Stage IV | 2/208 (1.0%) | 0/21 | 0/41 | 2/146 (1.4%) | n/a |
Including 30 patients with Deleterious and Likely Deleterious mutations and one MUTYH compound heterozygote
Including patients with a VUS only or a single MUTYH variant
Including patients with no Deleterious Variants, Likely Deleterious Variants or VUSs
Comparisons were made using a two-tailed Fisher’s exact test; except for comparison of age, Penn II scores, and BOADICEA scores which used a two-tailed, type 2 Student’s t-test. Comparisons were also run for Mutation positive versus Mutation and VUS negative (excluding the VUS positive patients) and all p-values were consistent. NS = not significant.
Other includes individuals of Asian descent (4), Hispanic/Latinos (6), and individuals reporting more than one race (2).
Any malignancy, excluding non-melanoma skin cancer.
Figure 1. Variants identified by multiplex panel testing of 278 patients with early onset breast cancer.
Germline DNA from 278 BRCA1/2 negative patients with early onset breast cancer (early-onset breast cancer) was isolated and subjected to massively parallel sequencing using a custom capture for the indicated genes in Bin A and Bin B. Sequencing data was analyzed with a custom bioinformatics pipeline and deleterious variants were called into classes (D = Deleterious, LD = Likely Deleterious, VUS = Variant of Uncertain Significance, LB = Likely Benign, and B = Benign). Inset: Proportion of patients self-reported as “White” or “Non-white” with deleterious or likely deleterious variants, VUSs only, or no reportable deleterious or likely deleterious variants or VUSs. The MUTYH heterozygous carriers included three patients heterozygous for a deleterious variant and three patients heterozygous for a VUS.
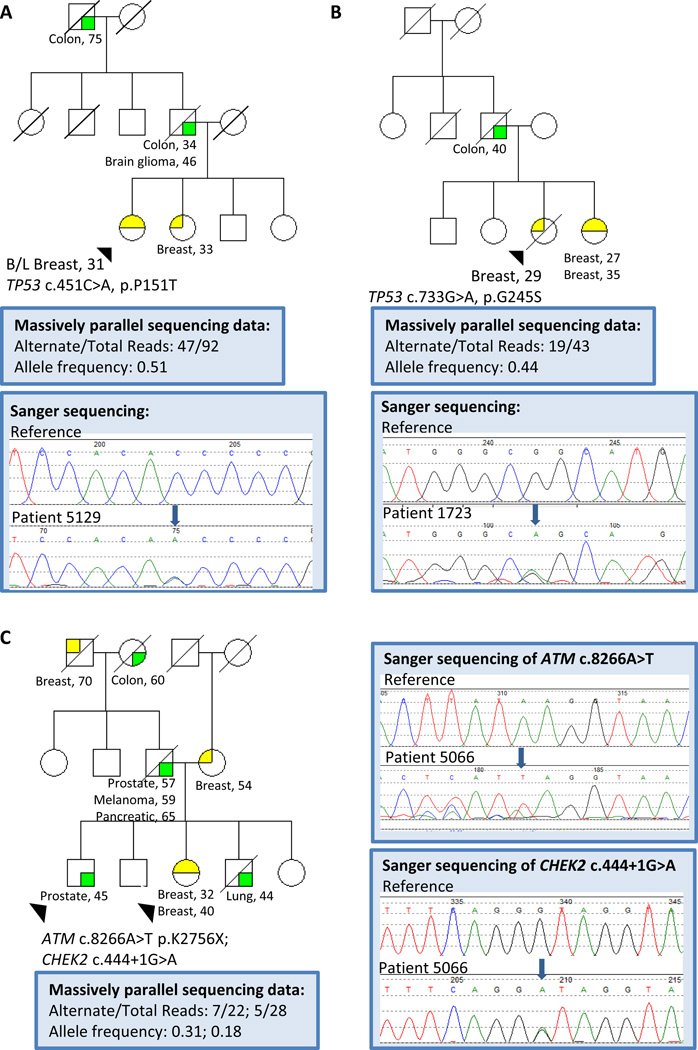
Seven patients were identified to have deleterious or likely deleterious variants in a high penetrance cancer susceptibility gene (Figure 1, Table 2). Two patients were found to carry a known pathogenic TP53 mutation (Figure 2a, b). Two patients, including one African American, were found to carry likely deleterious variants in TP53. One patient was identified to have a large genomic rearrangement deleting exon 5 of MSH2 leading to an in-frame deletion of 65 amino acids of the DNA interacting domain of MSH2. A patient with a history of both early-onset breast cancer and sarcoma was found to carry a known pathogenic missense mutation in CDKN2A. Finally, one patient, with a personal history of early onset colon cancer and two primary breast cancers, was found to be a compound heterozygote for a known pathogenic mutation and a likely deleterious variant in MUTYH.
Table 2.
Characteristics of patients with Deleterious or Likely Deleterious Variants
| Variant(s)a | Proband Cancer | Raceb and Family Historyc |
|---|---|---|
| TP53 c.451C>A, p.P151T (D) | Bilateral breast-31, ER-Her2+ | Race: W; M: Breast age>40, lymphoma; P: Breast age<40, colon ×2, brain |
|
TP53 c.733G>A, p.G245S (D) PALB2 c.94C>G, p.L32V (V) |
Breast-29, Unk | Race: U; Sib: Bilateral breast age<40; M: Breast age>40; P: colon |
| TP53 c.374C>T, p.T125M (LD) | Bilateral breast-30, DCIS | Race: A; M: None; P: Unknown |
| TP53 c.1000G>C, p.G334R (LD) | Breast-37, Unk; Breast-65,Unk |
Race: W/AJ; Sib: Colon; M: Breast age>40 ×3, leukemia, lymphoma, kidney, sarcoma, melanoma; P: colon |
|
CDKN2A c.104G>C, p.G35A (D) MSH6 c.3203G>A, p.R1068Q (V) |
Breast-38, Unk; Sarcoma-44 |
Race: W; M: Breast age>40, Melanoma; P: None |
| MSH2 del ex5 (LD) | Breast-39, ER+ Her2- | Race: W/AJ; M: None; P: thyroid, testicular |
|
MUTYH c.1187G>A, p.G396D (D); MUTYH c.281G>A, p.R94Q (LD) |
Colon-31; Breast-38, Unk; Breast-44, ER+Her2- | Race: W; M: Breast age<50, colon ×3, uterine; P: gallbladder |
| ATM c.8856delTC (D) | Breast-39, ER+ Her2+ | Race: W; M: None; P: pancreatic ×2, bladder, unknown gastrointestinal |
| ATM c.2282delCT (D) | Breast-39, DCIS | Race: A; M: Breast age>40 ×4; P: None |
| ATM c.6839delA (D) | Breast-34, DCIS | Race: W; M: Breast age<40 ×6, breast age>40 ×3, pancreatic, prostate, melanoma, brain; P: breast age>40 ×3, pancreatic |
| ATM c.7271T>G, p.V2424G (D) | Breast-29, ER+ Her2- | Race: A; M: Breast age>40; P: rectal, lung, brain ×2 |
| ATM 8774G>T, p.G2925V (LD) | Breast-31, ER+ Her2- | Race: W; M: Breast age>40 ×2, leukemia; P: None |
| ATM c.8155C>T, p.R2719C (LD) | Breast-38, ER+ Her2- | Race: A; M: Breast age>40; P: prostate |
| ATM c.8558C>G, p.T2853R (LD) | Breast-38, ER+ Her2- | Race: A; M: uterine; P: lung |
|
ATM c.8266A>T, p.K2756X (D) CHEK2 c.444+1G>A (D) |
Breast-32, Unk; Breast-40, ER+ Her2- |
Race: W; Sib: lung, prostate age 45; M: Breast age>40; P: prostate, melanoma, pancreatic, colon, breast age>40 |
| CHEK2 c.1100delC (D) | Breast-32, ER+ Her2- | Race: W; M: melanoma, breast age>40 ×2, colon ×3, uterine; P: Breast age<40×2 & age>40×2, brain |
| CHEK2 c.1100delC (D) | Breast-38, ER+ Her2- | Race: W; M: lung, thyroid; P: lung ×2 |
| CHEK2 c.1100delC (D) | Melanoma-30; Breast-34, Unk |
Race: W/AJ; M: Breast age<40 & age>40×3, prostate ×4; P: None |
| CHEK2 c.1555C>T, p.R519X (D) | Breast-37, ER+ | Race: W; M: Breast age>40, brain; P: None |
| CHEK2 c.444+1G>A (D) | Breast-32, ER-Her2+ | Race: W; P: Breast age>40, prostate; M: Breast age >40×2, leukemia, pancreatic, unknown gastrointestinal |
| CHEK2 c.470T>C, p.I157T (D) | Breast-36, Unk; Breast-49, ER+ Her2- |
Race: W; M: Breast age<40 & age>40 ×2, lung; P: breast age>40 |
| CHEK2 c.470T>C, p.I157T (D) | Breast-23, ER+ Her2+ | Race: W/AJ; M: Breast age>40, testicular, colon; P: none |
| CHEK2 c.349A>G, p.R117G (D) | Wilms-2; Breast-33, ER+ Her2+ | Race: W; M: None; P: prostate |
|
CHEK2 c.1283C>T, p.S428F (D) PMS2 c.944G>A, p.R315Q (V) |
Breast-38, ER+Her2+ | Race: W; M:adrenal, bladder, lung; P: None |
|
CHEK2 c.499G>A, p.G167R (LD) CHEK2 c.506T>C, p.F169Sd (LD) |
Breast-29, ER+ Her2- | Race: W/AJ; M: None; P: None; Sib (twin): breast age<40 |
| BARD1 c.1652C>G, p.S551X (D) | Breast-35, Unk; Breast-39, TNBC |
Race: W; M: None; P: Breast age>40 |
| BRIP1 c.2992delTT (D) | Breast-35, ER+; Bladder-44 |
Race: W/AJ; M: Breast age>40, colon, liver; P: Breast age>40, Lung |
| MRE11A c.1378G>T, p.E460X (D) | Breast-36, ER+ Her2+ | Race: W; M: Breast age<40, Breast age>40×2; P: lung |
| MRE11A c.1090C>T, p.R364X (D) | Breast-36, ER+ | Race: W; M: None; P: Breast age>40×3, uterine |
| RAD50 c.1252delTT (D) | Breast-31, ER+ Her2- | Race: A; M: Breast, Bone; P: None |
| NBN c.664T>C, p.F222L (LD) | Breast-37, Unk; Leukemia-39 |
Race: W; M: Breast age>40×2, P: melanoma, prostate, bladder, lymphoma |
D: deleterious variant, LD: likely deleterious variant, V: variant of unknown significance (VUS). The method of variant classification is described in the Methods section. Data supporting call for missense variants is provided in Supplementary Table 1.
W: White/Caucasian, A: African American, U: unknown; AJ: Ashkenazi Jewish descent
M: Cancers found on the maternal side, P: Cancers found on the paternal side; Sib: cancers found in siblings
The two CHEK2 mutations were shown to be in trans by analysis of 250 sequencing reads in IGV.
Figure 2. Representative family histories and sequencing data for three probands with identified mutations.
A. Patient 5129, TP53 c.451C>A, p.P151T found by massively parallel sequencing and confirmed by Sanger sequencing. B. Patient 1723, TP53 c.733G>A, p.G245S found by massively parallel sequencing and confirmed by Sanger sequencing. C. Patient 5066, ATM c.8266A>T p.K2756X and CHEK2 c.444+1G>A found by massively parallel sequencing and confirmed by Sanger sequencing in both the proband and her brother (arrows).
Twenty-four patients were found to have deleterious or likely deleterious variants in genes in which mutations have been associated with a moderate risk of breast cancer. The majority of deleterious or likely deleterious variants in moderate penetrance genes were found in ATM and CHEK2 (Figure 1, Table 2). Single deleterious or likely deleterious variants were found in ATM in seven patients and in CHEK2 in nine patients. One patient was found to carry deleterious variants in both ATM and CHEK2; of note both variants also were found in her brother with early onset prostate cancer (Figure 2c). In addition, one patient was found to carry two likely deleterious variants in trans in CHEK2. The remaining six patients had deleterious variants in MRE11A (2), BARD1 (1), BRIP1 (1), NBN (1), and RAD50 (1). Twenty-seven patients carried a VUS in a high penetrance cancer susceptibility gene, and three of those patients also had a deleterious or likely deleterious variant. Nine patients were found to have a single VUS in BRCA1 or BRCA2, three patients in TP53 and 12 patients in MLH1, MSH2, MSH6, or PMS2; no VUSs were found in CDH1, CDKN2A, STK11 or PTEN. Three additional patients each carried two VUSs in a high penetrance cancer susceptibility gene. Twenty-six patients were found to have VUSs in moderate penetrance cancer susceptibility genes, ATM, BRIP1, CHEK2, FAM175A, MRE11A, NBN, PALB2, RAD50, and RAD51C; no VUSs were found in BARD1. Finally, six patients carried a single deleterious variant or VUS in MUTYH (Figure 1). Three patients were heterozygous for the same known pathogenic MUTYH mutation and three were heterozygous for VUSs in MUTYH.
The proportion of patients identified to have a clinically reportable variant varied by race, such that 28% of self-reported white patients were found to have at least one reportable variant versus 37% of non-white patients (Figure 1, p=NS). The proportion of patients with a deleterious or likely deleterious variant did not vary significantly between white and non-white patients (13% versus 6%, p=NS). The proportion of non-white patients found to carry a VUS was statistically significantly higher than the proportion of white patients, 31% versus 15% (p=0.01). Of the 66 African Americans, 7.5% carried a deleterious or likely deleterious variant, which was not statistically significantly different than the proportion of white patients. Of the 27 Ashkenazi Jewish individuals, 22% were found to have a deleterious or likely deleterious variant, compared with 10% of the 234 non-Ashkenazi Jewish individuals (p=NS).
In comparison to deleterious or likely deleterious variant negative patients, there was a statistically significant increase in the rate of second primary malignancies (excluding non-melanoma skin cancers, Table 1, 19% vs 6%, p=0.02) in the deleterious or likely deleterious variant positive patients. In addition, there was a trend towards a higher rate of a bilineal family history of breast cancer in deleterious or likely deleterious variant positive versus negative patients (23% vs 11%, p=0.08). The Penn II BRCA1/2 prior probability score16 was statistically significantly higher (27% vs 19%, p=0.04) in deleterious or likely deleterious variant positive patients versus variant negative individuals, as was the BOADICEA17 score (29% vs 14%, p=0.005).
Only three of the 22 patients with deleterious or likely deleterious variants had ER- invasive breast cancer (Table 1, 14%), one had triple negative breast cancer (BARD1 p.S551X) and two had ER- Her2+ breast cancer (TP53 p.P151T and CHEK2 c.444+1A>G). In contrast, 33% of the patients with no deleterious or likely deleterious variant (+/− a VUS) had ER- invasive breast cancer (p=0.09). Seven of the 20 patients (35%) with a deleterious or likely deleterious variant had Her2+ breast cancer versus 26% of the patients with no deleterious or likely deleterious variant (+/− a VUS, p=NS). Finally, deleterious or likely deleterious variants were found in 13% of the patients with DCIS, 11% of the 116 patients with node positive invasive cancer, and 11% of the 130 patients with node negative invasive breast cancer. The stage distribution was similar between deleterious or likely deleterious variant positive versus negative patients.
DISCUSSION
Using massively parallel sequencing for 22 genes previously associated with cancer susceptibility, we found that 31% of BRCA1/2 negative patients with early-onset breast cancer and no family history of ovarian cancer have a clinically reportable variant, of which one-third were deleterious or likely deleterious variants. However, clinical guidelines exist for the management of cancer risk in only 2.5% of the patients, those found to have deleterious or likely deleterious variants in TP53, CDKN2A, MSH2, and the MUTYH double heterozygote. Currently, there are no standard of care clinical guidelines for the management of cancer risk in the 10% of women with single mutations in a moderate penetrance cancer susceptibility gene and MUTYH. Even greater clinical uncertainty exists for the 19% of patients who were found to carry VUSs.
Multiplex panel MPS-based mutation detection accurately identifies patients with mutations in genes leading to inherited cancer predisposition18 and has been used successfully to identify the spectrum of variants in single populations of patients with colon, ovarian and uterine cancers19–21. Recently, studies have reported findings using multiplex panels in heterogeneous groups of BRCA1/2 negative patients, either in randomly selected22,23 or consecutive24 patients from high risk genetics clinics or in all patient samples submitted to commercial testing laboratories23,25. Excluding monoallelic MUTYH carriers as the associated cancer risks are controversial26, these studies of predominantly white individuals found that between 3.4–9.5% of BRCA1/2 negative patients carried deleterious or likely deleterious variants in panel genes22–25. We found a deleterious or likely deleterious variant rate of 11% using a custom 22-gene panel in a well-characterized group of 278 early-onset breast cancer patients, including 66 African Americans, consistent with an increased likelihood of finding cancer susceptibility mutations in a younger, affected patient population. We found that 2.2% were heterozygous MUTYH carriers, similar to the LaDuca study rate of 1.7%25 and the reported population carrier frequency of MUTYH mutations of 1.1% (range 0–2%)27.
Our variant classification algorithm found a 19% VUS rate in the early-onset breast cancer patients using a pipeline integrating multiple data sources. Kurian et al. used only two in silico variant calling programs and population frequency data to analyze variants and reported a much higher 88% VUS rate. Our VUS rate is consistent with that in LaDuca et al. of 20% identified using Ambry’s proprietary variant calling program, although lower than Tung et al of 42% using Myriad’s variant calling method23. Given that VUSs cause confusion and anxiety for both patients and practitioners, incorporating various data sources to support calls and exploring novel variant classification methods will be increasingly necessary going forward.
In our study, we found that seven patients (2.5%) carried clinically reportable variants in TP53. Regarding the four individuals with TP53 deleterious or likely deleterious variants, two had family histories meeting Chompret criteria, one was diagnosed at age 30 with bilateral breast cancer and one had a family history of late-onset sarcoma and multiple late-onset bilateral breast cancer cases; all were ascertained prior to 2007. No mutations were found in the genes associated with other well characterized cancer susceptibility syndromes, PTEN, STK11, and CDH1. Many of the patients in this study population were reviewed in a genetics conference at a tertiary care institution where there is high index of awareness for these phenotypes, and patients with known mutations in these genes were excluded from the present study. Their mutation rates may differ in unselected populations.
With regard to other high risk cancer susceptibility genes, one patient with a family history of melanoma was found to have a mutation in CDKN2A; excess breast cancer has been described in families with CDKN2A mutations28. One patient was found to have a likely deleterious variant in MSH2 and one patient was a compound heterozygote for a MUTYH pathogenic mutation and a likely deleterious variant; the breast cancer risks associated with mutations in MUTYH and the mismatch repair genes such as MSH2 is controversial29,30. It is possible that these mutations did not contribute to the development of breast cancer in these individuals. Further study of the breast cancer risks associated with these gene mutations is needed. These data highlight the importance of determining the clinical management of individuals identified to have mutations by multiplex panel testing in genes not classically associated with the patient’s phenotype or pedigree.
Regarding moderate risk breast cancer susceptibility genes, we found ATM mutations in 2.9% (n=8), CHEK2 founder mutations (1100delC, I157T and c.444+1G>A) in 2.5% (n=7), and other CHEK2 mutations in 1.4% (n=4) of patients. In addition, we found two patients with MRE11A mutations and single patients with mutations in BARD1, BRIP1, NBN, and RAD50, respectively. Interestingly, we did not identify any patients with PALB2 or RAD51C mutations. It is possible that the ethnic diversity of our population (28% non-white) is responsible for the variability in mutation frequency between ours and other studies31–35. Our study demonstrates that mutations in individual moderate penetrance genes outside of ATM and CHEK2 are likely very infrequent in patients with early-onset breast cancer.
There are a number of important limitations to our study. Our study design excluded individuals with a personal or family history of ovarian cancer and it is possible that such early-onset breast cancer patients will have a different spectrum of mutations. Our study also did not include genes recently proposed to contribute to breast cancer susceptibility such as BLM36, FANCC36, and XRCC237 or ovarian cancer susceptibility such as RAD51D38, and mutations in these genes could be present in our study population. Massively parallel sequencing approaches have limitations in the identification of large genomic rearrangements and therefore these types of variants could still be present in our patient population. Finally, as the majority of patients in the study had a family history of breast cancer and were ascertained through two health systems and affiliated hospitals, our findings may not be generalizable to patients with early-onset breast cancer ascertained through population based studies.
Overall, our results suggest that at least 11% of BRCA1/2 negative patients with early-onset breast cancer may have a causative mutation in high or moderate penetrance genes found on multiplex panel testing. A higher incidence of other malignancies may occur in early-onset breast cancer patients with these mutations, and further study of these risks in larger populations could allow for more rational decision making regarding cancer screening and medical and/or surgical preventive treatments for these patients3, for example prophylactic contralateral mastectomy at the time of a breast cancer diagnosis. In addition, it is now understood that the tumors in BRCA1/2 carriers show increased sensitivity to PARP inhibitors and platinum agents due to synthetic lethality39. Given that many of the other cancer susceptibility genes studied here also play a role in double stranded DNA repair, it is possible that tumors of carriers of some of these other gene mutations may also show increased sensitivity to these agents40.
Although our sample size was too limited to define the breast and non-breast cancer risks for family members of individuals with mutations in moderate penetrance genes, the Penn II and BOADICEA model prior probability scores were statistically significantly higher in deleterious or likely deleterious variant positive patients and this may reflect the stronger family histories of breast and/or other cancers in patients with deleterious mutations. Additional studies are needed to determine if true negative family members of those with mutations in the genes studied here can be counseled that they are at population risk for breast and other gene specific cancers, as is the case for BRCA1/23.
Our results highlight the critical need for large consortia to delineate the expected mutation rates, penetrance, and associated cancer risks for moderate risk genes found on cancer susceptibility genetic testing panels in well-defined clinical populations, keeping in mind the relatively lower penetrance of some of these mutations and the possibility for segregation of multiple risk alleles. In addition, consortia will be needed to pool data to study and develop clinical recommendations for patients carrying these mutations and their family members.
Supplementary Material
Acknowledgements
The authors would like to acknowledge the funding sources for this work, the Department of Defense, National Institutes of Health, American Association for Cancer Research, Breast Cancer Research Foundation, Rooney Family Foundation, Basser Center for BRCA Research at the University of Pennsylvania, MacDonald Cancer Risk Evaluation Program, Susan G Komen Foundation, and CURE (Commonwealth Universal Research Enhancement) Program (KLN). The authors would like to express their sincere gratitude to the patients who have provided samples for this research and the health care professionals who recruited patients included in this study.
Footnotes
Disclaimers: The Pennsylvania Department of Health specifically disclaims responsibility for any analyses, interpretations or conclusions. Views and opinions of, and endorsements by the authors do not reflect those of the US Army or the Department of Defense. All work contained in this manuscript is original.
This study was presented in part at the AACR Annual Meeting 2013 and as an oral presentation in a Clinical Science Symposium at the ASCO Annual Meeting 2014.
References
- 1.Sundquist M, Thorstenson S, Brudin L, Wingren S, Nordenskjold B. Incidence and prognosis in early onset breast cancer. Breast. 2002;11:30–35. doi: 10.1054/brst.2001.0358. [DOI] [PubMed] [Google Scholar]
- 2.Daly MB, Axilbund JE, Buys S, et al. Genetic/familial high-risk assessment: breast and ovarian. J Natl Compr Canc Netw. 2010;8:562–594. doi: 10.6004/jnccn.2010.0043. [DOI] [PubMed] [Google Scholar]
- 3.Maxwell KN, Domchek SM. Cancer treatment according to BRCA1 and BRCA2 mutations. Nat Rev Clin Oncol. 2012;9:520–528. doi: 10.1038/nrclinonc.2012.123. [DOI] [PubMed] [Google Scholar]
- 4.Maxwell KN, Domcheck SM. Familial Breast Cancer Risk. Current Breast Cancer Reports. 2013;5:170–182. [Google Scholar]
- 5.Shendure J, Aiden EL. The expanding scope of DNA sequencing. Nat Biotechnol. 2012;30:1084–1094. doi: 10.1038/nbt.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stadler ZK, Schrader KA, Vijai J, Robson ME, Offit K. Cancer genomics and inherited risk. J Clin Oncol. 2014;32:687–698. doi: 10.1200/JCO.2013.49.7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biesecker LG, Burke W, Kohane I, Plon SE, Zimmern R. Next-generation sequencing in the clinic: are we ready? Nat Rev Genet. 2012;13:818–824. doi: 10.1038/nrg3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Domchek SM, Bradbury A, Garber JE, Offit K, Robson ME. Multiplex genetic testing for cancer susceptibility: out on the high wire without a net? J Clin Oncol. 2013;31:1267–1270. doi: 10.1200/JCO.2012.46.9403. [DOI] [PubMed] [Google Scholar]
- 9.Zheng Y, Ogundiran TO, Falusi AG, et al. Fine mapping of breast cancer genome-wide association studies loci in women of African ancestry identifies novel susceptibility markers. Carcinogenesis. 2013;34:1520–1528. doi: 10.1093/carcin/bgt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pabinger S, Dander A, Fischer M, et al. A survey of tools for variant analysis of next-generation genome sequencing data. Briefings in bioinformatics. 2014;15:256–278. doi: 10.1093/bib/bbs086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fromer M, Moran JL, Chambert K, et al. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet. 2012;91:597–607. doi: 10.1016/j.ajhg.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woolfe A, Mullikin JC, Elnitski L. Genomic features defining exonic variants that modulate splicing. Genome biology. 2010;11:R20. doi: 10.1186/gb-2010-11-2-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tavtigian SV, Oefner PJ, Babikyan D, et al. Rare, evolutionarily unlikely missense substitutions in ATM confer increased risk of breast cancer. Am J Hum Genet. 2009;85:427–446. doi: 10.1016/j.ajhg.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez-Perez A, Lopez-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet. 2011;88:440–449. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindor NM, Johnson KJ, Harvey H, et al. Predicting BRCA1 and BRCA2 gene mutation carriers: comparison of PENN II model to previous study. Fam Cancer. 2010;9:495–502. doi: 10.1007/s10689-010-9348-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee AJ, Cunningham AP, Kuchenbaecker KB, Mavaddat N, Easton DF, Antoniou AC. BOADICEA breast cancer risk prediction model: updates to cancer incidences, tumour pathology and web interface. Br J Cancer. 2014;110:535–545. doi: 10.1038/bjc.2013.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walsh T, Lee MK, Casadei S, et al. Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc Natl Acad Sci U S A. 2010;107:12629–12633. doi: 10.1073/pnas.1007983107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pennington KP, Walsh T, Lee M, et al. BRCA1, TP53, and CHEK2 germline mutations in uterine serous carcinoma. Cancer. 2013;119:332–338. doi: 10.1002/cncr.27720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cragun D, Radford C, Dolinsky J, Caldwell M, Chao E, Pal T. Panel-Based Testing for Inherited Colorectal Cancer: A descriptive study of clinical testing performed by a U.S. Laboratory. Clin Genet. 2014 doi: 10.1111/cge.12359. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walsh T, Casadei S, Lee MK, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:18032–18037. doi: 10.1073/pnas.1115052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurian AW, Hare EE, Mills MA, et al. Clinical Evaluation of a Multiple-Gene Sequencing Panel for Hereditary Cancer Risk Assessment. J Clin Oncol. 2014;32:2001–2009. doi: 10.1200/JCO.2013.53.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tung N, Battelli C, Allen B, et al. Frequency of mutations in individuals with breast cancer referred for BRCA1 and BRCA2 testing using next-generation sequencing with a 25-gene panel. Cancer. 2014 doi: 10.1002/cncr.29010. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 24.Castéra L, Krieger S, Rousselin A, et al. Next-generation sequencing for the diagnosis of hereditary breast and ovarian cancer using genomic capture targeting multiple candidate gene. Eur J Hum Genet. 2014 doi: 10.1038/ejhg.2014.16. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laduca H, Stuenkel AJ, Dolinsky JS, et al. Utilization of multigene panels in hereditary cancer predisposition testing: analysis of more than 2,000 patients. Genet Med. 2014 doi: 10.1038/gim.2014.40. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Win AK, Dowty JG, Cleary SP, et al. Risk of colorectal cancer for carriers of mutations in MUTYH, with and without a family history of cancer. Gastroenterology. 2014;146:1208–1211. e1201–e1205. doi: 10.1053/j.gastro.2014.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peterlongo P, Mitra N, Chuai S, et al. Colorectal cancer risk in individuals with biallelic or monoallelic mutations of MYH. Int J Cancer. 2005;114:505–507. doi: 10.1002/ijc.20767. [DOI] [PubMed] [Google Scholar]
- 28.Borg A, Sandberg T, Nilsson K, et al. High frequency of multiple melanomas and breast and pancreas carcinomas in CDKN2A mutation-positive melanoma families. J Natl Cancer Inst. 2000;92:1260–1266. doi: 10.1093/jnci/92.15.1260. [DOI] [PubMed] [Google Scholar]
- 29.Win AK, Lindor NM, Jenkins MA. Risk of breast cancer in Lynch syndrome: a systematic review. Breast Cancer Res. 2013;15:R27. doi: 10.1186/bcr3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beiner ME, Zhang WW, Zhang S, Gallinger S, Sun P, Narod SA. Mutations of the MYH gene do not substantially contribute to the risk of breast cancer. Breast Cancer Res Treat. 2009;114:575–578. doi: 10.1007/s10549-008-0042-1. [DOI] [PubMed] [Google Scholar]
- 31.FitzGerald MG, Bean JM, Hegde SR, et al. Heterozygous ATM mutations do not contribute to early onset of breast cancer. Nat Genet. 1997;15:307–310. doi: 10.1038/ng0397-307. [DOI] [PubMed] [Google Scholar]
- 32.Cao AY, Huang J, Hu Z, et al. Mutation analysis of BRIP1/BACH1 in BRCA1/BRCA2 negative Chinese women with early onset breast cancer or affected relatives. Breast Cancer Res Treat. 2009;115:51–55. doi: 10.1007/s10549-008-0052-z. [DOI] [PubMed] [Google Scholar]
- 33.Cao AY, Huang J, Hu Z, et al. The prevalence of PALB2 germline mutations in BRCA1/BRCA2 negative Chinese women with early onset breast cancer or affected relatives. Breast Cancer Res Treat. 2009;114:457–462. doi: 10.1007/s10549-008-0036-z. [DOI] [PubMed] [Google Scholar]
- 34.Ding D, Zhang Y, He X, Meng W, Ma W, Zheng W. Frequency of the CHEK2 1100delC mutation among women with early-onset and bilateral breast cancer. Breast Cancer Res. 2012;14:401. doi: 10.1186/bcr3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foulkes WD, Ghadirian P, Akbari MR, et al. Identification of a novel truncating PALB2 mutation and analysis of its contribution to early-onset breast cancer in French-Canadian women. Breast Cancer Res. 2007;9:R83. doi: 10.1186/bcr1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson ER, Doyle MA, Ryland GL, et al. Exome sequencing identifies rare deleterious mutations in DNA repair genes FANCC and BLM as potential breast cancer susceptibility alleles. PLoS Genet. 2012;8:e1002894. doi: 10.1371/journal.pgen.1002894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park DJ, Lesueur F, Nguyen-Dumont T, et al. Rare mutations in XRCC2 increase the risk of breast cancer. Am J Hum Genet. 2012;90:734–739. doi: 10.1016/j.ajhg.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson ER, Rowley SM, Sawyer S, et al. Analysis of RAD51D in ovarian cancer patients and families with a history of ovarian or breast cancer. PLoS One. 2013;8:e54772. doi: 10.1371/journal.pone.0054772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 40.Pennington KP, Walsh T, Harrell MI, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res. 2014;20:764–775. doi: 10.1158/1078-0432.CCR-13-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.