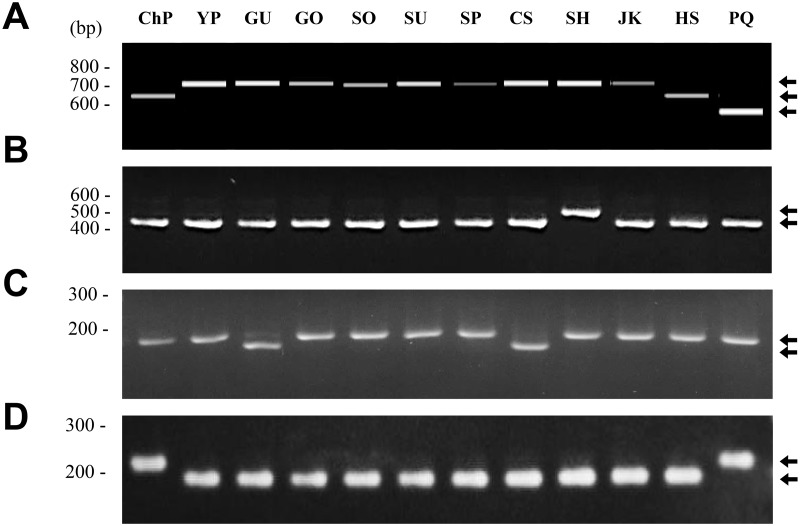
Fig 5. Validation of SNP and InDel polymorphic sites.
(A, B) PCR analysis of InDel regions in ycf1 (A) using primer set pgycf1 and trnUUC-trnGGU (B) using primer set pgcp137 (Table 3). (C) SNP analysis using dCAPS primers, pgcpd02, designed for the SNP site in the rpoC2 gene (Table 3). ScaI digestion of the amplicon produced a digested fragment except in GU and CS. (D) SNP analysis with dCAPS primers, pgcpd01, designed for the SNP site in rpoC1 gene (Table 3). XbaI digestion of the amplicon produced a digested fragment except in ChP and PQ. Abbreviated cultivar names (defined in Table 1) are denoted on the gels. PQ and M denote P. quinquefolius and 100-bp DNA ladder, respectively.