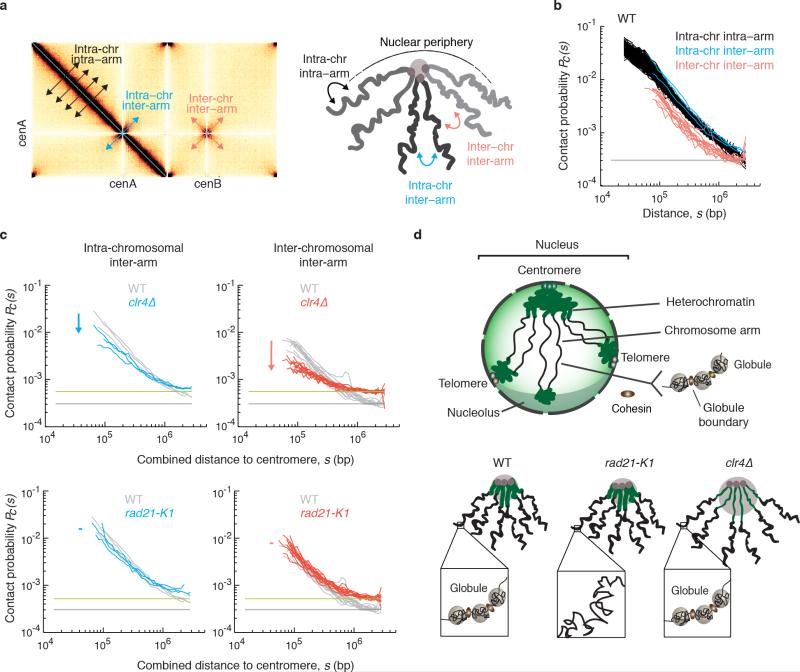
Extended Data Figure 10. Effects of heterochromatin and cohesin on centromere proximal arm interactions, and a model showing their distinct effects on S. pombe genome organization.
a, Centromere proximal regions considered for intra-chromosomal and inter-chromosomal arm interaction Pc(s) are shown on a heatmap (left). Diagram showing intra- and inter-arm interactions within a chromosome, and inter-arm interactions between chromosomes in centromere proximal regions (right). Standard genomic distances were used to consider intra-arm contact probabilities. To consider inter-arm contacts within or between chromosomes, genomic distance was defined as the combined distance of two loci to the centromere, for regions at similar (<50kb) distance from their respective centromeres. b, Pc(s) plotted for inter-arm interactions within a chromosome and between different chromosomes in wild type cells. Inter-chromosomal inter-arm (red) Pc(s) falls below intra-chromosomal inter-arm (blue) and intra-arm (black), but have a similar rate of decay with distance. Note that inter-arm Pc(s) starts at a larger genomic distance, since centromere proximal bins were removed at the stage of bin-level filtering due to their low coverage. c, Pairwise comparisons of inter-arm Pc(s) of rad21-K1 and clr4Δ with wild type. In clr4Δ, both inter-arm Pc(s) are shifted lower, most notably for inter-chromosomal inter-arm contact probability. In rad21-K1, inter-arm scaling near centromere is similar to wild type. d, Model showing distinct roles of heterochromatin and cohesin-dependent globules in overall chromosome organization. In wild type cells, non-random organization consistent with a degree of chromosome territoriality was evident. These levels of organization may underlie genomic integrity, both independently and collectively, for example, by effectively preventing interaction between repetitive elements. The peripheral positioning of centromere and telomere clusters promotes a Rabl configuration. Heterochromatin reinforces this configuration by compacting centromere and telomere proximal regions, promoting strong interactions and aligning arms to facilitate proper genome architecture. A newly identified layer of “globules” bounded by high amounts of arm cohesin organizes chromosome arms. The formation of globules depends on arm cohesin. Unlike in wild type, globules on arms are disrupted in rad21-K1, whereas constraints at pericentromeric regions are maintained. In clr4Δ, pericentromeric regions are decompacted, but globules are not disrupted along arms.