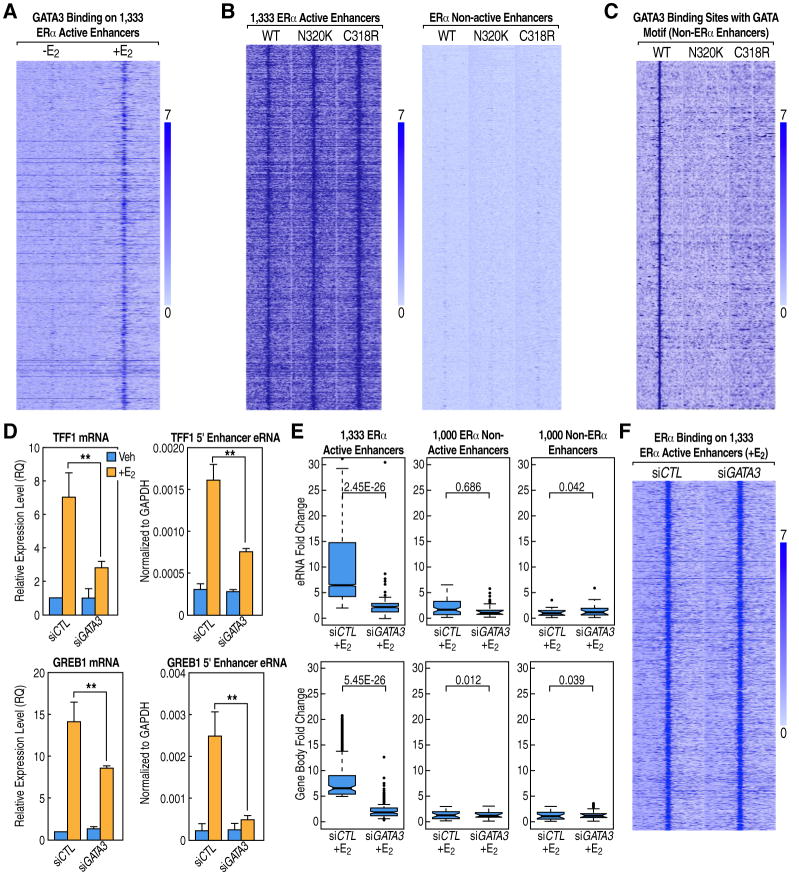
Figure 3. Trans-Bound GATA3 on ERα Active Enhancers Regulates ERα E2-Liganded Transcription Activation.
(A) Heatmap displaying GATA3 binding at the 1,333 ERα active enhancers is enhanced by E2.
(B) Heatmaps of ChIP-seq data for wild-type and two DNA-binding mutants of GATA3 (+E2) show the binding of GATA3 to these ERα active enhancers is not dependent on its DNA-binding ability. There is no binding of either wild-type or mutants GATA3 to ERα non-active enhancers.
(C) Heatmap of ChIP-seq data for wild-type and two DNA-binding mutants of GATA3 (+E2) shows the binding of GATA3 to these non-ERα enhancers that contain the GATA motif requires its DNA-binding ability.
(D) Knockdown GATA3 affects ERα-dependent activation of eRNA transcription and coding gene expression for GREB1 and TFF1 genes. Mean ± SEM based on three independent qPCR experiments (** P<0.01).
(E) GRO-seq boxplots showing that GATA3 is required for the E2-liganded activation of ERα active enhancers and their coding gene targets.
(F) Heatmap showing that knockdown of GATA3 does not affect ERα binding at the 1,333 active enhancers.
See also Figure S4.