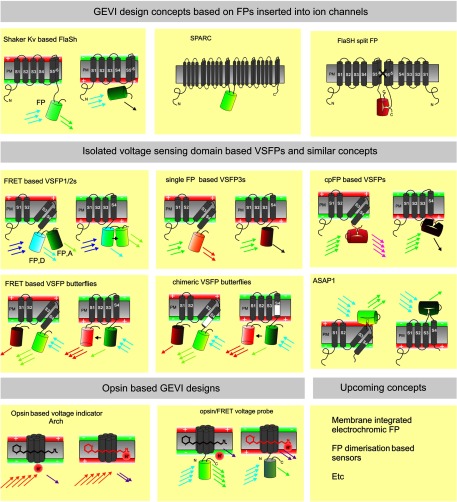
Fig. 1.
Overview of genetically encoded voltage indicator (GEVI) molecular designs: Upper row shows the GEVIs based on the insertion of fluorescent proteins (FPs) (depicted as barrels) into complete ion channel proteins with segments (e.g., S1–S6) that cross the plasma membrane (PM). In the FlaSh-type voltage indicator, a FP is fused into the C-terminal portion of a Shaker potassium channel subunit. Tetramers of subunits form a channel structure which is made nonconducting by a point mutation. Modulation of FlaSh fluorescence is triggered by voltage-dependent rearrangements, probably corresponding to channel C-type inactivation. Middle panels show the GEVIs based on isolated voltage-sensing domains. In Förster resonance energy transfer (FRET)-based voltage-sensitive probes of the voltage-sensitive fluorescent protein (VSFP1/2) type, the voltage-sensor domain, consisting of four segments (S1–S4), is fused to a pair of FPs (FP, D: FRET donor; FP, A: FRET acceptor). A change in membrane potential induces a rearrangement of the two FPs that is optically reported as a change in the ratio between donor and acceptor fluorescence. Single FP and circularly permuted (cp) FP probes of the VSFP3 family are monochromatic. In FRET-based voltage-sensitive probes of the VSFP Butterfly family, the voltage-sensor domain is sandwiched between two FPs. Lower panels show the GEVIs based on opsins. A change in membrane potential induces increased fluorescence of the retinal molecule. The microbial rhodopsin-based voltage indicator Arch shows an increased fluorescence of the retinal molecule when the membrane potential is increased. In opsin/FRET probes, a FP in quenched by retinal in a voltage-dependent manner.