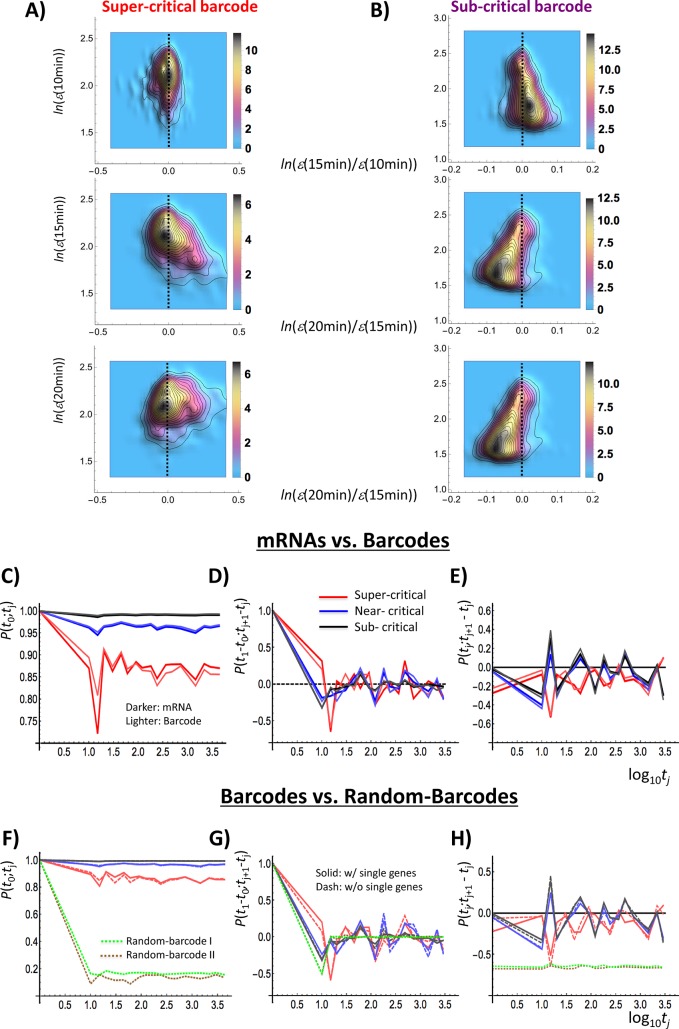
Fig 9. Synchronization of barcode genes on chromosomes with mRNA coherent expression dynamics.
Panel A): super-critical state, Panel B): sub-critical state for barcode genes. The panels present the probability density functions of barcode genes on the regulatory space (first row: x: change in the natural logarithm of expression at 10–15 min; y: natural logarithm of expression at 10 min); second row: x: change at 15–20 min; y: at 15 min; third row: x: change at 15–20 min; y: at 20 min), showing a similar opposite oscillation between sub-critical and super-critical states in mRNA expression dynamics (Fig 6A). Panels C-E): mRNAs versus barcode genes. Panels F-H): a variety of barcode genes. Temporal Pearson correlations for sub-critical as well as near-critical barcode genes confirm the ON-OFF synchronization with mRNA expression dynamics, the same in terms of stochasticity and coherent oscillation (coherent stochastic oscillation: CSO; D: stochasticity; E: coherent oscillation), whereas super-critical barcodes reveal the opposite phase of CSO, showing similar temporal trend to critical states of mRNA (mRNAs (dark colors): subcritical: black; near-critical: blue; super-critical: red; barcodes: the corresponding lighter colors). Panels C & F: Pearson correlation, P(t 0;t j); Panels D & G: P(t 1-t 0;t j+1 -t j), and Panels E & H: P(t j;t j+1−t j). Temporal Pearson correlations (F-H) confirm that barcodes without single gene (multiple genes: dashed lines) clearly follow similar trends of temporal correlations of the whole barcodes of critical states (solid lines), and thus similar correlation response to the mRNA dynamics. Correlation trends of barcodes are shown to be clearly distinct from random-barcodes: one (green dotted line: random-barcode I) for average value of randomly selected barcodes (n = 200) out of the whole barcodes combining critical states with 100 repeats–forming Gaussian distribution, and another (brown dotted line: random-barcode II) for barcode genes (N = 3130) of randomly mixed critical states, where genes on chromosome are randomly selected, and for each selected barcode, the number of elements is assigned randomly from 1 to 4 neighboring genes.