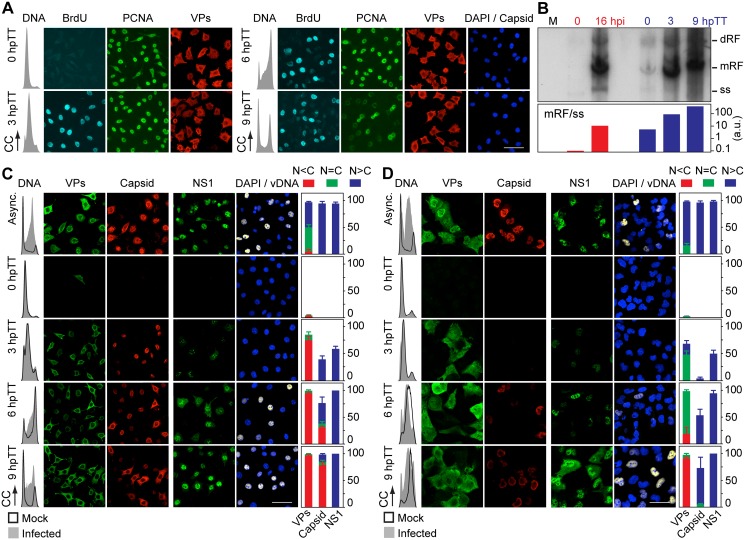
Fig 4. Stressing cellular DNA synthesis uncouples parvovirus genome replication from nuclear capsid assembly.
A. Cellular DNA synthesis dependence of VPs subcellular distribution and assembly in transfected MFs. The figure shows the subcellular distribution of BrdU incorporation, PCNA, VPs, and Capsid in MF-VPs analyzed by confocal microscopy at the indicated hours post-release of the double thymidine block (hpTT). Left: histograms of cell cycle analysis by flow cytometric determination of DNA content. B. Stressing cellular DNA synthesis hampers the packaging of parvovirus genome. Southern-blot analysis (performed with a full-lenght 32P-MVM probe as described [42]) of viral replication and maturation in growing (hpi) and thymidine synchronized (hpTT) infected MFs. Low molecular weight DNA isolated from 105, or from 5x105 (in 0 hpTT and 3 hpTT) cells, was loaded per lane. M, mock infection. mRF and dRF, monomeric and dimeric viral replicative intermediates, respectively; ss, single-stranded viral genomes. Below, mRF/ss ratio of synthesis measured by densitometry. a.u, arbitrary units. DNA synthesis stress hampers parvovirus capsid assembly in infected MFs (C) and HFs (D). The subcellular distribution of VPs, assembled capsid (Capsid), NS1, and viral replication analyzed by FISH-hybridization (vDNA), is shown by confocal microscopy in synchronously infected mouse and human fibroblasts at the indicated hpTT. Async: non-synchronous cultures. DNA (left columns): cell cycle phases at the respective time post-release analyzed by cytometry (histograms: blank for mock, grey-filled for infected). Right bars: percentages with standard errors of subcellular distribution of the viral antigens. Scale bar in IF is 50 μm.