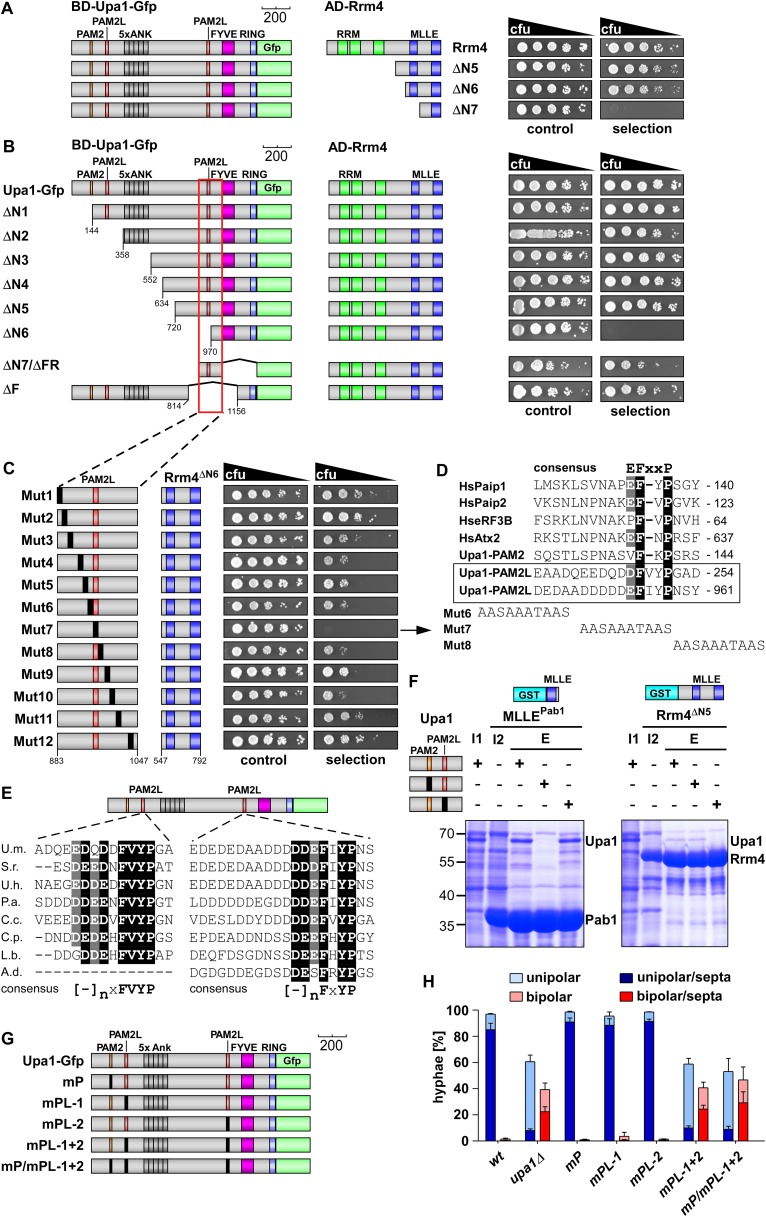
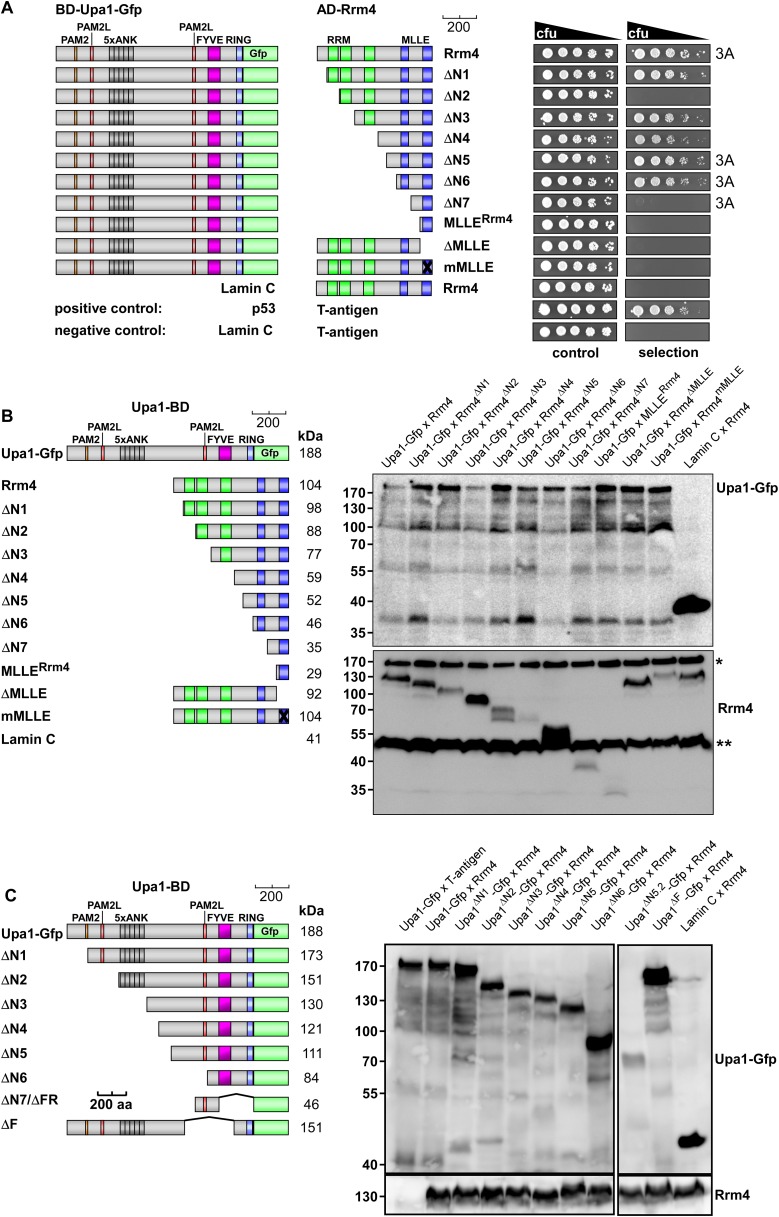
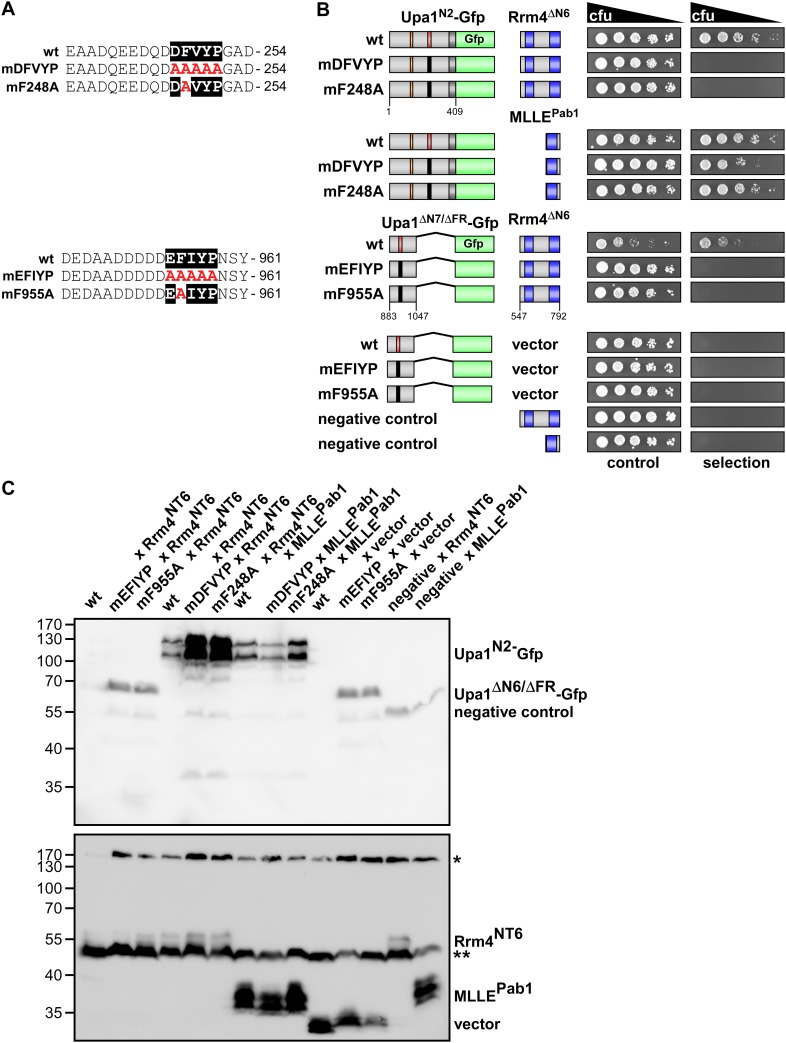
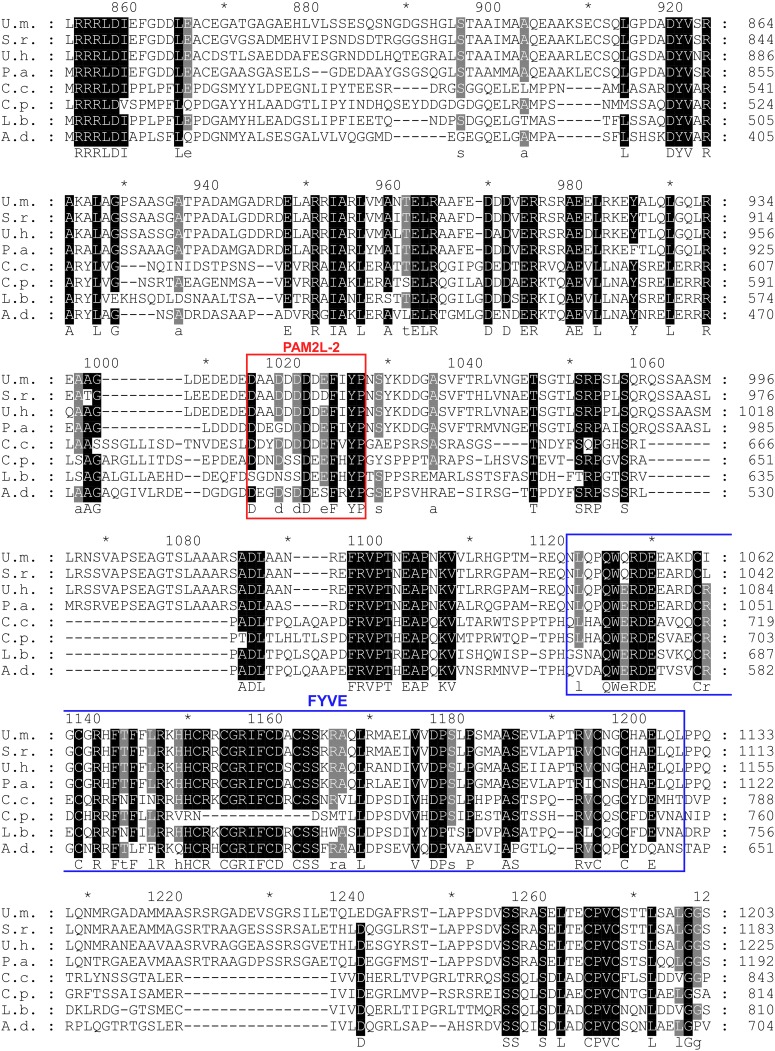
Figure 3. Upa1 contains two PAM2L motives for interaction with Rrm4.
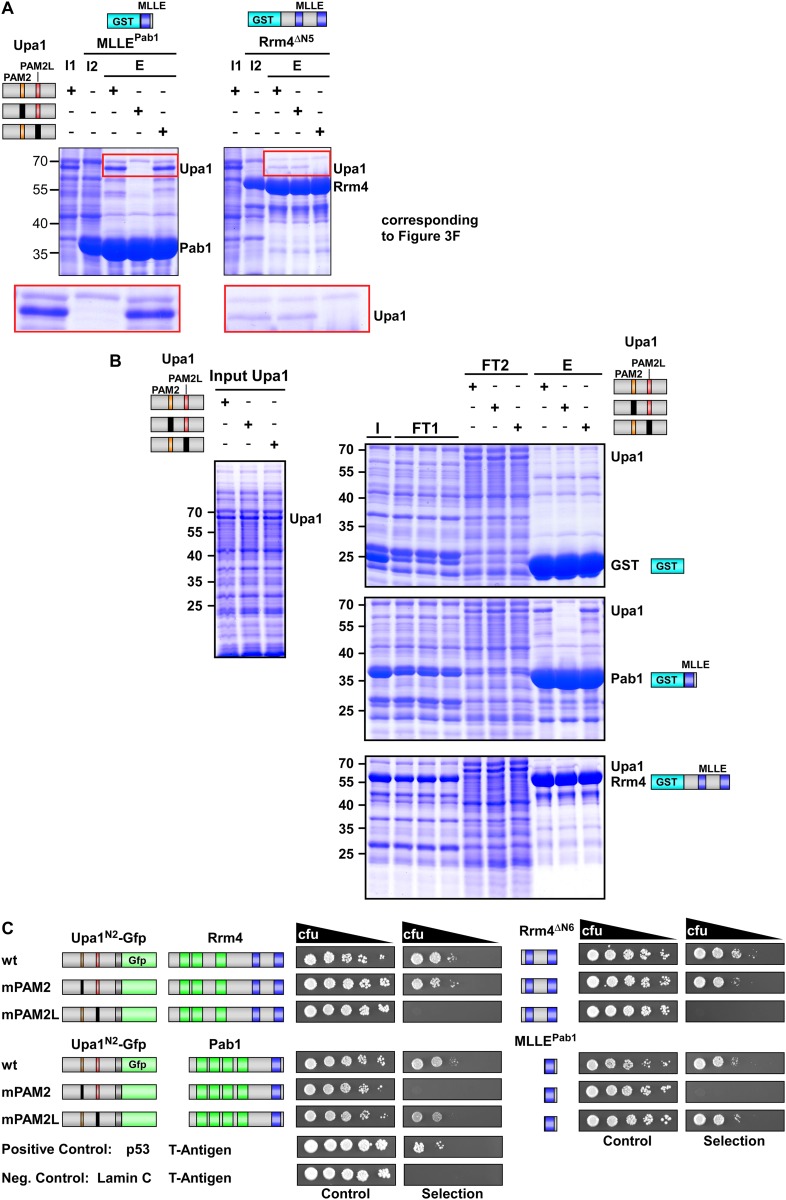
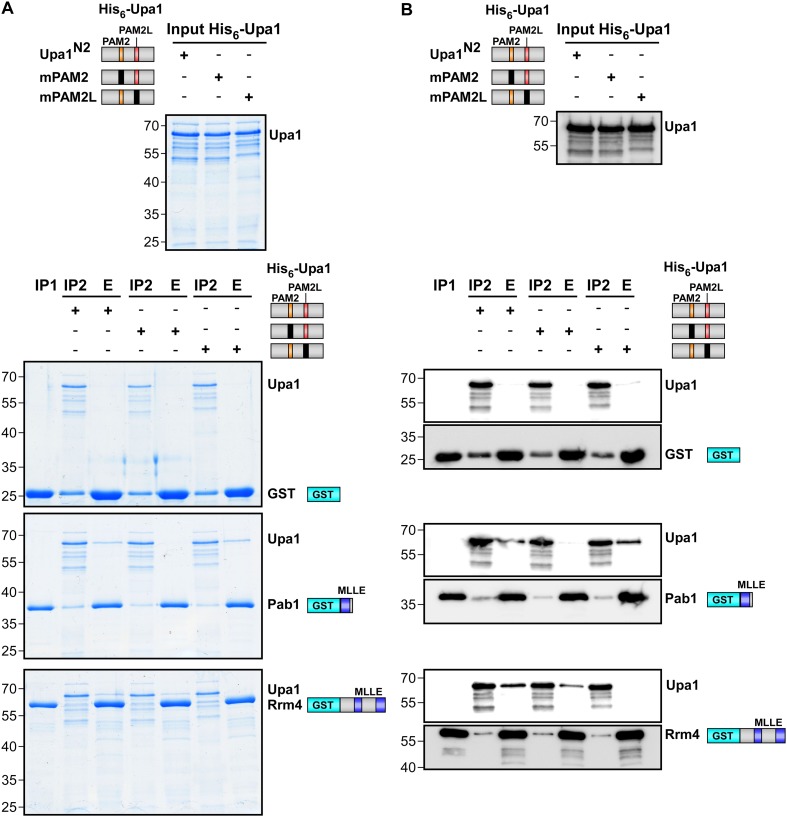
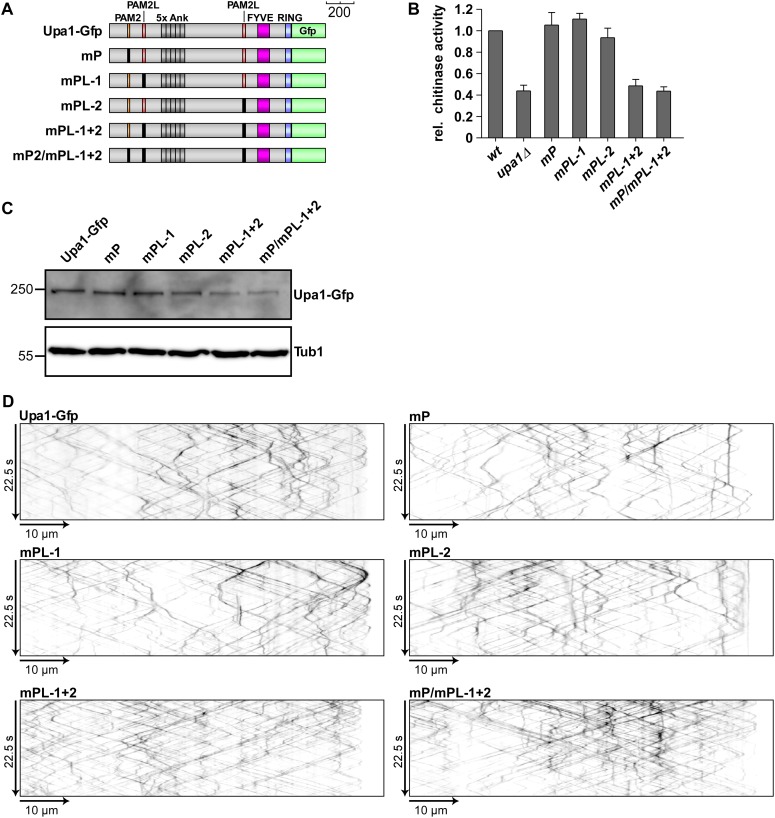
(A) Two-hybrid analysis with schematic representation of variants tested (left) and growth plates (right). Yeast cultures were serially diluted 1:5 (decreasing cfu) and spotted on respective growth plates assaying for reporter gene expression. (B) Two-hybrid analysis as in (A). Red rectangle indicates minimal region in Upa1 interacting with Rrm4. (C) Two-hybrid analysis as in (A). Upa1 region identified in (B) was analysed by linker scanning mutagenesis (mutations indicated as black bar, Mut1-12). (D) Comparison of PAM2 and PAM2L sequences as in Figure 1B. Note, the second PAM2L motif was only mutated in Mut7 (E) PAM2L sequences of Upa1 compared to related sequences from basidiomycetes (U.m., Ustilago maydis UMAG_12183 / XP_758247.1; S.r., Sporisorium reilianum, accession number sr13323 / CBQ72642.1; U.h., Ustilago hordei accession number UHOR_03,485 / CCF52210.1; P.a. Pseudozyma antarctica GAK65366.1; C.c. Coprinopsis cinerea CC1G_00,427 / XP_001837291.2; C.p. Coniophora putanea XP_007767511.1; L.b. Laccaria bicolor XP_001876756.1; A.d. Auricularia delicate XP_007337909.1). (F) GST co-purification experiments with components expressed in E. coli N-terminal His6-tagged versions of Upa1, Upa1mP, and Upa1mPL (amino acids 1–363) were expressed to the same level (first input lane, I1; see Figure 3—figure supplement 5B). MLLE domains of Pab1 or Rrm4 (MLLEPab1 or Rrm4ΔN5, respectively) were expressed as GST fusion proteins (second input lane, I2). After GST affinity chromatography proteins were eluted (lanes marked with "E"). Interaction studies were performed with whole protein extracts from E. coli to demonstrate specific binding. (G) Schematic representation of Upa1 variants carrying mutations (black boxes) in the PAM2 and PAM2L regions. (H) Percentage of hyphae (8 h.p.i.): unipolarity, bipolarity, and septum formation was quantified (error bars, s.e.m.; n = 3 independent experiments, >100 hyphae were counted per experiment; note that septum formation is given relative to the values of unipolar or bipolar hyphae set to 100%).