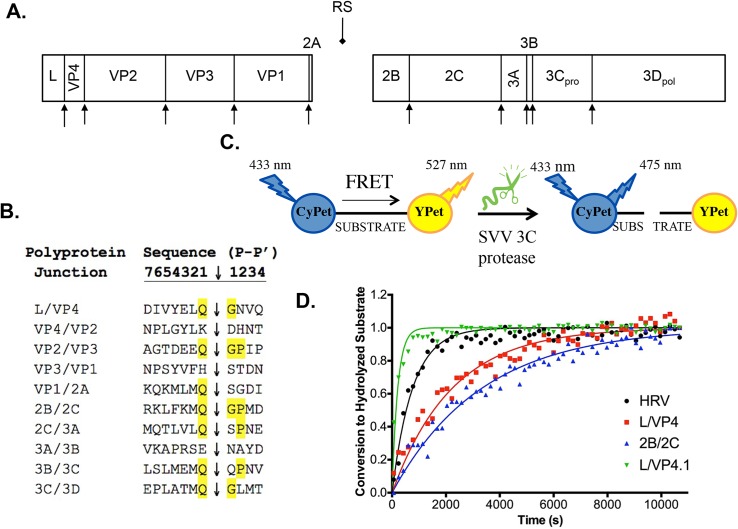
Fig 2. SVV-001 polyprotein and identification of efficient 3C protease substrates A.
The SVV-001 polyprotein is hypothesized to include twelve mature proteins based on sequence alignment with closely related viruses. Protein regions (drawn to scale) are presented with proposed 3Cpro cleavage sites (arrowheads) depicted. The 2A ribosome skipping sequence (RS; diamondhead arrow) is also shown. B. Sequence alignment of proposed 3Cpro cleavage sites. Sites are named based on the two mature proteins flanking these sites. The presumed scissile bond is depicted with an arrowhead with the consensus Q↓GP cleavage sequence amino acids highlighted. C. Schematic of FRET substrate construction and kinetic assays. Substrates are cloned between two fluorescent proteins, CyPET and YPET. These molecules exhibit FRET when in close proximity; therefore, there will be a high level of emission at 527 nm (YPET) and lower emission at 475 nm (CyPET). As the 3C protease cleaves and releases YPET from proximity to CyPET, the amount of FRET will decrease observed as an increase in CyPET emission (475 nm) and a decrease in YPET emission (527 nm). D. Conversion of FRET substrates by purified HRV or SVV-001 3Cpro over time. Data points represent the average of three replicates at each time point. The HRV substrate was used as a positive control. L/VP4 and 2B/2C are endogenous substrates. L/VP4.1 (P2’ N→P substitution) is a further optimized version of the endogenous L/VP4 substrate. Lines of the same color correspond to curve fits from GraphPad. Error bars on data points were removed for figure clarity.