Fig 3. Optimization of the L/VP4 Substrate A.
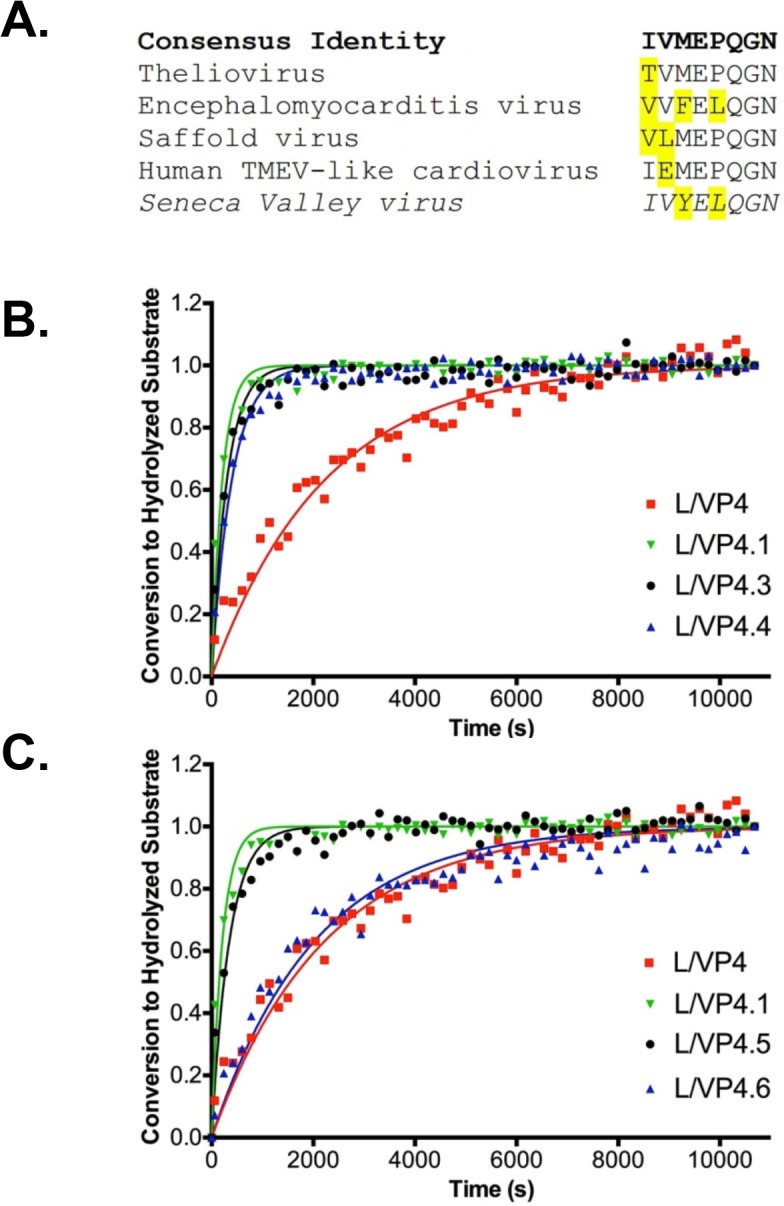
Alignment of SVV L/VP4 cleavage site (shown in italics) with L/VP4 junction sequences of closely related cardioviruses. The amino acids in each sequence that diverge from the consensus sequence (shown in bold) are highlighted. B. Conversion of modified L/VP4.1 FRET substrates by purified SVV-001 3Cpro over time. Data points represent the average of three replicates at each time point. The endogenous L/VP4 substrate and optimized L/VP4.1 substrate are shown for reference. L/VP4.3 and L/VP4.4 are P4 Y→F substitution and P4 Y→M substitution substrates, respectively. Lines of the same color correspond to curve fits from GraphPad. Error bars on data points were removed for figure clarity. C. Conversion of truncated L/VP4.1 FRET substrates by purified SVV-001 3Cpro over time. Data points represent the average of three replicates at each time point. The endogenous L/VP4 substrate and optimized L/VP4.1 substrate are shown for comparison. L/VP4.5 and L/VP4.6 are P6 and P5/P6 truncations of L/VP4.1, respectively. Lines of the same color correspond to curve fits from GraphPad. Error bars on data points were removed for figure clarity.
