Fig 4. SVV-001 3Cpro substrates are cleaved in the context of a cellular infection.
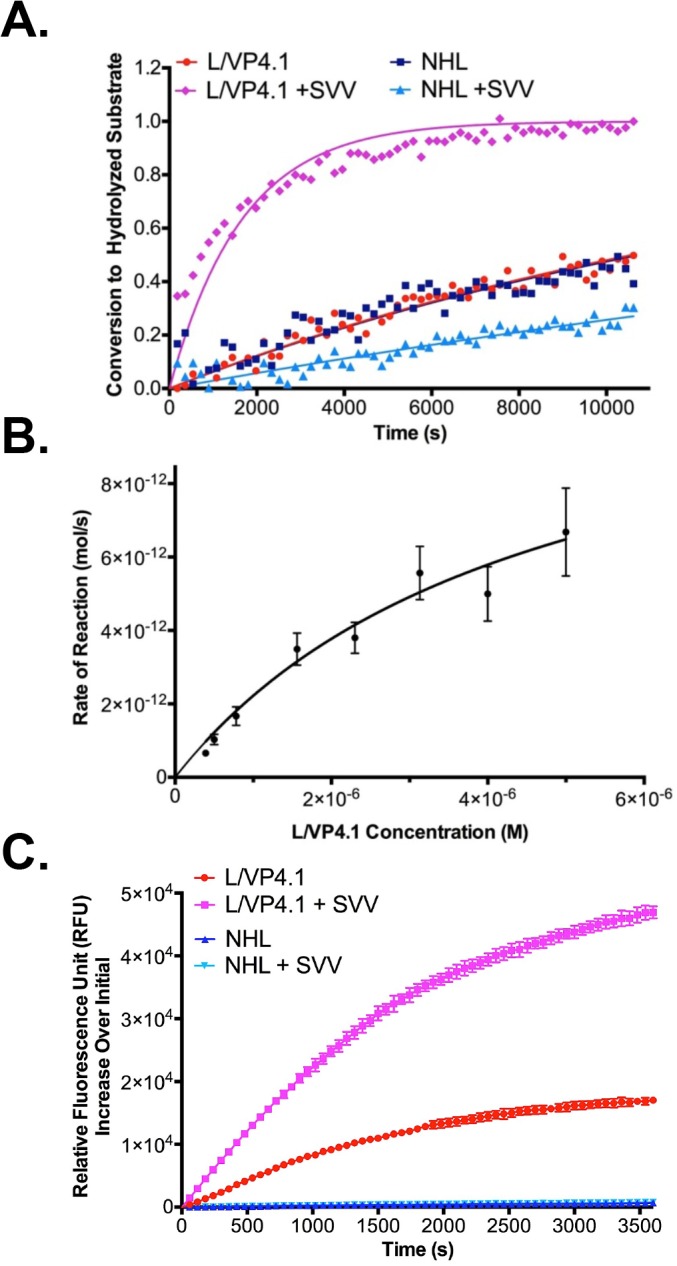
A. Conversion of FRET substrates by SVV-001 3Cpro produced by a cellular SVV infection of permissive SCLC line, NCI-H446. Data points represent the average of three replicates at each time point. The L/VP4.1 FP substrate incubated with uninfected and infected cells, using similar incubations of NHL FP substrate as a negative control. Lines of the same color correspond to curve fits from GraphPad. Error bars on data points were removed for figure clarity. B. Initial reaction rates of L/VP4.1 peptide cleavage by recombinant SVV-001 3Cpro. Data points represent the initial rate of reaction at each concentration of L/VP4.1 peptide calculated from three replicate experiments. Data points were fit to a Michaelis-Menten nonlinear regression from GraphPad and the kinetic constants determined by the curve fit were reported. Standard deviation values of the kinetics constants were calculated by the GraphPad software and propagated through second order rate constant calculations. C. Proteolysis of CPQ2/5-FAM peptides by native SVV-001 3Cpro in a cellular assay with NCI-H446. Data points represent the average relative fluorescence units (RFUs) increase of three replicates at each time point relative to fluorescence at time zero. The L/VP4.1 FQ peptide was incubated with uninfected and infected cells, using similar incubations of NHL FQ peptide as a negative control. Lines of the same color correspond to connecting line between points.
