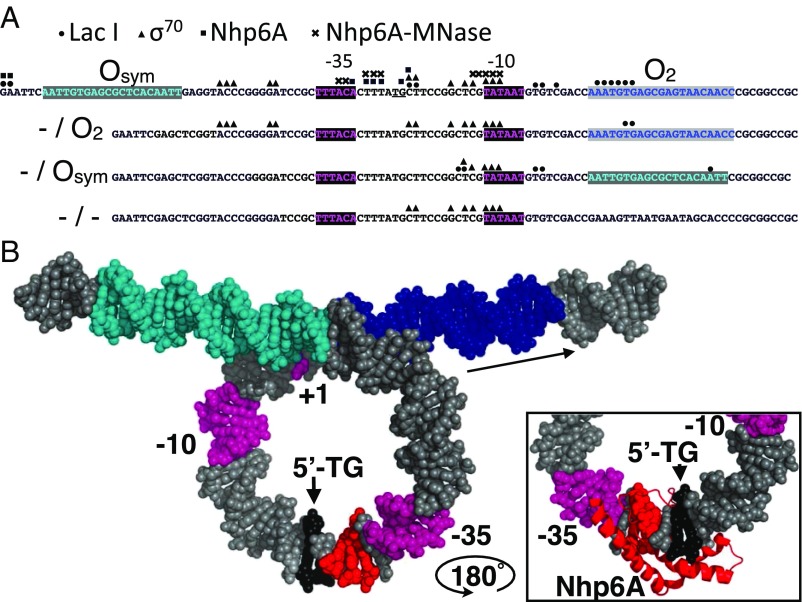
Fig. 4.
Summary of high-resolution protein binding data and model. (A) Four tested lac promoter constructs with the indicated lac operators and promoter elements showing protein binding sites identified by ChIP-exo-LMPCR (LacI, circles; σ70, triangles; Nhp6A, squares near TG/CA dinucleotide, underlined), and by ChEC-LMPCR (Nhp6A-MNase, crosses). (B) Model of LacI loop showing operators (cyan and blue), promoter elements (magenta), and Nhp6A cleavage sites (red) identified by ChEC-LMPCR near the TG/CA dinucleotide (black) proposed as the kinked binding site. (Inset) Illustration of plausible intercalation of Nhp6A methionine 29 at the TG/CA dinucleotide based on [PDB ID code 1J5N (25)] after rotation of the complex by 180° about a vertical axis.