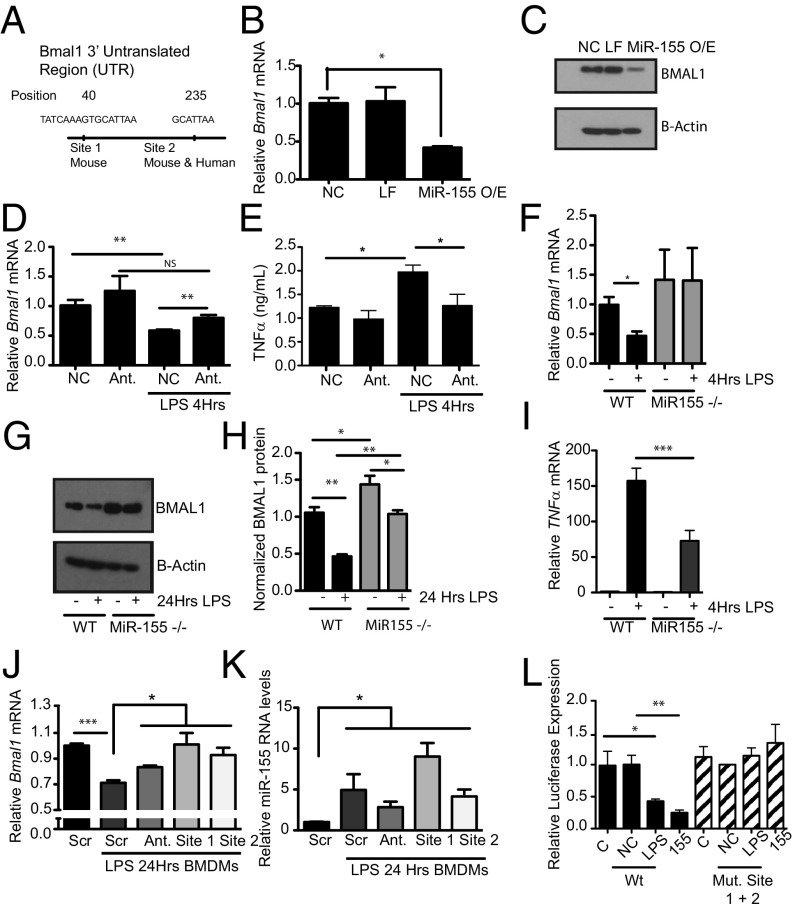
Fig. 3.
Bmal1 is repressed by the microRNA MiR-155 under basal and LPS conditions. (A) Schematic of Bmal1 3′ UTR illustrating position of the two miR-155–binding sites identified with the software TargetScan. iBMDMs transfected with a miR-155 mimic and analyzed for expression of (B) Bmal1 mRNA and (C) protein (n = 3). NC, negative control for mimic; LF, Lipofectamine; MiR-155O/E, overexpression of MiR-155 mimic. iBMDMs transfected with either a negative control for antagomir (NC) or an antagomir (Ant) to miR-155, treated with LPS (100 ng/mL), and analyzed for expression of (D) Bmal1 and (E) TNFα levels by ELISA (n = 3). WT or miR-155−/− BMDMs treated with LPS (100 ng/mL) for the indicated time and analyzed for expression of (F) Bmal1 and (G) protein levels by immunoblot. (H) Densitometry values of immunoblots from G and Fig. S4 (n = 3–4), and (I) Tnfα mRNA (n = 3). BMDMs transfected with a scrambled control morpholino (Scr), MiR-155 antagomir (Ant.) morpholino against the MiR-155 site in Bmal1 at position 40 (site 1), and morpholino against the MiR-155 site in Bmal1 at position 235 (site 2) were treated with LPS (100 ng/mL) for 24 h and analyzed for expression of (J) Bmal1 and (K) MiR-155. (L) Luciferase reporter activity from Bmal1 3′ UTR construct with LPS induction (100 ng/mL) or overexpression of MiR-155 mimic with wild-type luciferase construct or double (Mut. Site 1 + 2) mutations of miR-155–binding sites (n = 3). C, control; NC, negative control for mimic; 155, overexpression of miR-155 mimic. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001.