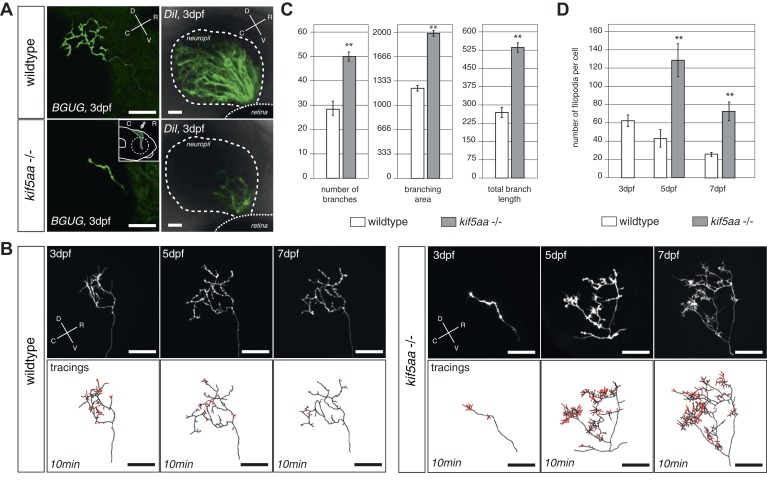
Figure 3. RGC axons in kif5aa mutants show a delayed ingrowth into the optic tectum and grow larger arbors at later stages.
(A) Single membrane-GFP expressing RGC axons from the Tg(BGUG) transgene (left panel) and DiI injections into the contralateral retina of 3 dpf old wild-type and kif5aa mutant embryos (right panel) illustrate the delay of tectal innervation in mutants. Scale bars = 20 μm. The schematic in the lower left panel illustrates the perspective chosen for image acquisition (indicated by an arrow). D = dorsal, V = ventral, R = rostral, C = caudal. (B) Upper panel, left: Axonal arbor of a single wild-type RGC at 3, 5, and 7 dpf. Lower panel, left: Tracings of an axonal arbor at time point zero. In red: Overlay of filopodia formed and retracted within 10 min (1 frame/2 min). Scale bars = 20 μm. Upper panel, right: Axonal arbor of a single kif5aa mutant RGC axonal arbor at 3, 5, and 7 dpf. Lower panel, right: Tracing of an axonal arbor at time point zero. In red: Overlay of filopodia formed and retracted within 10 min (1 frame/2 min). Scale bars = 20 μm. (C) Kif5aa mutant RGC axons grow significantly more branches, cover a larger area of the optic tectum with their arbors (in μm2) and grow longer arbors (in μm) than wild-type cells at 7 dpf (n = 10, p < 0.01). Scale bars = 20 μm. For quantification, only branches stable within 10 min of image acquisition were selected. (D) Quantification of filopodia numbers formed and retracted within 10 min per cell at 3, 5, and 7 dpf. While wild-type RGC arbors form most filopodia at 3 dpf and reduce this rate constantly until 7 dpf, kif5aa mutant RGC axons grow almost three times more filopodia at 5 dpf (n = 4, p < 0.01). At 7 dpf, the rate is still more as double as high as for their wild-type counterparts (n = 4, p < 0.01).