Abstract
Background
Recent studies have demonstrated that the clock gene PER1 regulates various tumor-related genes. Abnormal expressions and circadian rhythm alterations of PER1 are closely related to carcinogenesis. However, the dynamic circadian variations of PER1 and tumor-related genes at different stages of carcinogenesis remain unknown. This study was conducted to investigate the daily rhythm variation of PER1 and expression of tumor-related genes VEGF, KI67, C-MYC, and P53 in different stages of carcinogenesis.
Materials and methods
Dimethylbenzanthracene was used to establish a golden hamster model of buccal mucosa carcinogenesis. Hamsters with normal buccal mucosa, precancerous lesion, and cancerous lesion were sacrificed at six different time points during a 24-hour period of a day. Pathological examination was conducted using routine hematoxylin and eosin staining. PER1, VEGF, KI67, C-MYC, and P53 mRNAs were detected by real-time reverse transcriptase polymerase chain reaction, and a cosinor analysis was applied to analyze the daily rhythm.
Results
PER1, VEGF, C-MYC, and P53 mRNA exhibited daily rhythmic expression in three carcinogenesis stages, and KI67 mRNA exhibited daily rhythmic expression in the normal and precancerous stages. The daily rhythmic expression of KI67 was not observed in cancerous stages. The mesor and amplitude of PER1 and P53 mRNA expression decreased upon the development of cancer (P<0.05), whereas the mesor and amplitude of VEGF, KI67, and C-MYC mRNA increased upon the development of cancer (P<0.05). Compared with the normal tissues, the acrophases of PER1, VEGF, and C-MYC mRNA occurred earlier, whereas the acrophases of P53 and KI67 mRNA lagged remarkably in the precancerous lesions. In the cancer stage, the acrophases of VEGF and C-MYC mRNA occurred earlier and later, respectively, compared with the normal stage.
Conclusion
Variations in the daily rhythm characteristics of the clock gene PER1 and tumor-related genes VEGF, KI67, C-MYC, and P53 correlate with the development of cancer. Additional studies might provide new insights and methods to explore carcinogenic mechanisms and cancer treatment.
Keywords: circadian rhythm, clock gene, PER1, tumor, carcinogenesis
Introduction
“Circadian” is a Latin word that means “about a day”.1 Living organisms on earth have evolved with an endogenous timing system, specifically the circadian clock.1–3 Current research has demonstrated that numerous vital physiological processes that occur in mammals in vivo, such as sleeping and waking, hormone secretion, and immune activity, exhibit a circadian fluctuation of approximately 24 hours.1,4–7 The generation of a circadian rhythm in vivo depends on circadian variations in clock genes expression. At least 14 known core clock genes have been reported, including PER1, PER2, PER3, CRY1, CRY2, TIM, CKIε, CLOCK, BMAL1, RORs, REV-ERBs, NPAS2, DEC1, and DEC2.8–11 These clock genes are located in the suprachiasmatic nuclei of the anterior hypothalamus and probably in all peripheral tissue cells.12–14 The former is called the central circadian clock, and the latter is the peripheral circadian clock.15,16 Circadian rhythms in the central circadian and peripheral circadian clocks are generated by the interaction of positive and negative transcription–translation feedback loops at the molecular level. The transcriptional regulators CLOCK and BMAL1 form CLOCK/BMAL1 heterodimers, and the heterodimers subsequently bind to the E-box of the PER (PER1, PER2, and PER3) and CRY (CRY1 and CRY2) genes to activate the expression of these genes in the nuclei. These processes are the main components of the positive feedback circadian rhythm loop.17,18 PER and CRY proteins form heterodimers that translocate from the cytoplasm to the nucleus, where they interfere with CLOCK/BMAL1 heterodimer formation and inhibit the expressions of PER and CRY, thereby forming a negative feedback loop.11,19,20 The clock genes generate and sustain rhythmic expressions in a 24-hour period via these feedback loops.2,12 The peripheral circadian clock can be regulated and influenced by the central circadian clock; meanwhile, the peripheral circadian clock can also generate and sustain a 24-hour circadian rhythm independently.2,16 Approximately 5%–10% of the genes of the mammal genome are regulated by clock genes,3,21 which exhibit 24-hour periodic expression, and these genes are known as clock-controlled genes (CCGs).12,22 Importantly, the circadian expression of clock genes and CCGs maintains a high level of synchronization and orderliness for normal physiological activities.23,24
Current reports have demonstrated that the PER1 gene is an important clock gene that exhibits circadian rhythms in normal cells.25,26 PER1 is a cancer suppressor gene that is downregulated in various solid tumor cells, including prostatic cancer, melanoma, and oral squamous cell carcinoma.8,27,28 Many important cancer-related genes are CCGs; PER1 is an important clock gene that regulates many cancer-related genes. Gery et al reported that PER1 over-expression in human colon cancer cells (HCT116) increased P53 and C-MYC expression.29 Soták et al demonstrated that reduced PER1 mRNA expression in mice colorectal tumors promoted the upregulation of C-MYC and downregulation of P21.15
Current studies have demonstrated that the clock gene PER1 regulates numerous downstream cancer-related genes to influence cell proliferation and apoptosis as well as tumor angiogenesis; thus, PER1 is closely related to the occurrence and development of carcinoma.15,29,30 Carcinogenesis is a process of pathological change involving multiple genes and steps. The dynamic circadian rhythm variations (including the expressions of mesor, amplitude, and acrophase) of the clock gene PER1 and cancer-related CCGs involved in the stages of carcinogenesis and the development of carcinoma are unclear. The daily profiles of the expression of clock gene PER1 and cancer-related genes VEGF, KI67, C-MYC, and P53 during three stages of carcinogenesis in a golden hamster model of buccal mucosa carcinoma were detected in this study; this research provides new concepts for further research on the mechanism of carcinogenesis and the development of cancer as well as the development of cancer therapies based on the recovery of circadian rhythms.
Materials and methods
Experimental animals
Ninety male-specific pathogen-free Syrian golden hamsters (6–7 weeks, 90–120 g) were purchased from Vital River Company (Beijing, People’s Republic of China). All experimental procedures were approved by the Experimental Animal Use and Management Committee in Experimental Animal Research Institute, Chongqing Medical University.
Establishment of a Syrian golden hamster model of buccal mucosa carcinoma by dimethylbenzanthracene
Ninety golden hamsters were randomly housed in separate cages (five hamsters per cage). The golden hamsters were synchronized for 3 weeks under 12-hour light/12-hour dark cycles (at a temperature of 24°C±1°C and humidity of 60%±10%) before the study. The hamsters’ bedding, food, and water were sterilized. In the 12-hour light/12-hour dark cycles, the time was described as hours after light onset (HALO). For example, “0 HALO” was denoted as the time to turn on the light, whereas “12 HALO” was denoted as the time to turn off the light. On the last day of the third week, five hamsters were sacrificed by cervical dislocation at each time point, including 4 HALO, 8 HALO, 12 HALO, 16 HALO, 20 HALO, and 24 HALO during a 24-hour period, and normal left buccal mucosa was simultaneously obtained from the normal group. The remaining 60 hamsters were housed under a 12-hour light/12-hour dark cycle. The hamsters were maintained in a self-made box, and their mouths were opened and painted with a 0.5% dimethylbenzanthracene acetone solution (Sigma, USA) with a No 5 flat brush on the left buccal mucosa every Monday, Wednesday, and Friday. On the last day of the sixth and 14th week after dimethylbenzanthracene painting, five hamsters were sacrificed by cervical dislocation at each time point, including 4 HALO, 8 HALO, 12 HALO, 16 HALO, 20 HALO, and 24 HALO during a 24-hour period; the left buccal diseased tissue was obtained simultaneously. The tissues obtained in the sixth week were included in the precancerous lesions group, whereas tissues obtained in the 14th week were included in the cancer group. Then, each tissue sample was divided into two parts. One portion was fixed in 4% paraformaldehyde, dehydrated, and embedded in paraffin blocks, whereas the other portion was snap-frozen in liquid nitrogen.
Pathological examination using HE staining
We obtained 4 μm thick sections from each paraffin block, and the hematoxylin and eosin (HE)-stained sections were prepared for observation under an optical microscope (Olympus BX51, Japan). Classification of histological pathology was accomplished according to the classification standard of WHO Collaborating Centre for Oral Precancerous Lesions.31
Real-time RT-PCR
Real-time reverse transcriptase polymerase chain reaction (RT-PCR) was performed according to the manufacturer’s protocol. First, the total RNA was isolated using RNAiso Plus (Takara, Japan). Second, the cDNA was synthesized from 10 μL of total RNA using a Prime Script RT reagent kit (Takara). Finally, RT-PCR was performed on a real-time PCR machine (CFX96TM Real-Time PCR Detection System; BioRad, USA) in a 25 μL final volume containing 12.5 μL of 2× SYBR Premix Ex TaqTM II (Takara), 8.5 μL of RNAse-free water, 1 μL of 0.4 μmol/L forward primer and reverse primer, and 2 μL of 50 ng/μL cDNA. Specific primers for β-actin (a housekeeping gene that served as an internal control), PER1, VEGF, KI67, C-MYC, and P53 were designed using Oligo 17.0 software. The primer sequences are listed in Table 1. The following RT-PCR cycling parameters were used: 1.5 minutes at 95°C followed by 40 rounds of 10 seconds at 95°C and 30 seconds at 60°C. The data were acquired as the threshold cycle (Ct) value. The relative mRNA expression values of PER1, VEGF, KI67, C-MYC, and P53 were calculated using the 2−ΔΔCt method. Each sample was performed in triplicate to ensure the accuracy of the data.
Table 1.
Primers used for real-time PCR amplification of gene expression
| Gene | Primer | Primer sequence (5′ to 3′) |
|---|---|---|
| PER1 | Forward | AGCAACAGCCACGGTTCTC |
| Reverse | CAGTCCACACAAGCCATCAC | |
| VEGF | Forward | GACTTTGTTTGTCCAAGATCC |
| Reverse | TCACATCTGCAAGTACGTTCG | |
| KI67 | Forward | TGTATCCTTTGGTGGTCGTCTA |
| Reverse | GCTGGAGTGTGAGTGGTGAG | |
| C-MYC | Forward | GCCTACATCCTGTCCATCCA |
| Reverse | TCACGCACCAGAGTTTCG | |
| P53 | Forward | ATGCCGAATACCTGGATGAC |
| Reverse | GCGTGATGATGGTGAGGATA | |
| β-actin | Forward | GGCAGGCAAAGGTTACTCTG |
| Reverse | TGGTGACAGGTGGACAAGAT |
Abbreviation: PCR, polymerase chain reaction.
Statistical analysis
One-way analysis of variance and Student–Newman–Keuls test were used to analyze the differences in the mRNA expression of each gene among six time points and the differences in the mesor and amplitude of each gene in three groups with SPSS 17.0 statistical software (IBM Corporation, Armonk, NY, USA). Mean and standard deviation were used to describe the results, and a P-value of <0.05 was considered statistically significant. The daily rhythm was assessed by a single cosine test using Time Series Analysis Cosinor 6.3 software (Expert Soft Technologie Laboratory of Applied Statistics and BioMedical Computing, Richelieu, France). A value of P<0.05 indicated that the expression of the target gene showed a daily rhythm. A cosine-fitted curve was generated with SigmaPlot 10.0. The daily rhythm was characterized by the mesor, amplitude, and acrophase. The mesor is the mean value of all the statistics in a circadian fluctuation within a 24-hour period, the amplitude indicates the maximum degree above or below the mesor of the circadian fluctuation, and the acrophase represents the time point when the circadian fluctuation achieved the peak value.
Results
Pathological examination using HE staining
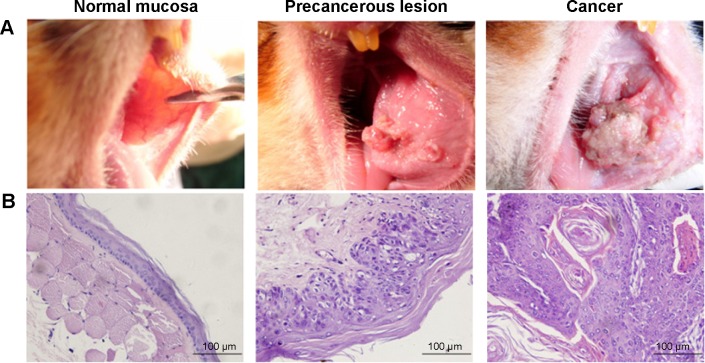
The pathological examination conducted using HE staining indicates that the 30 cases in the normal group exhibited normal buccal mucosa tissue. Among the 30 cases in the precancerous lesions group, 25 displayed moderate atypical hyperplasia, three exhibited mild atypical hyperplasia, and two exhibited severe atypical hyperplasia. The 30 cases in the cancer group exhibited squamous carcinoma tissue (Figure 1).
Figure 1.
The golden hamster buccal mucosa carcinogenesis model.
Notes: (A) Images of the golden hamster buccal mucosa from various carcinogenesis stages. (B) Pathological slice photos (HE staining, ×400) from various carcinogenesis stages.
Abbreviation: HE, hematoxylin and eosin.
Daily rhythm variations of the clock gene PER1 mRNA in various stages of buccal mucosa carcinogenesis
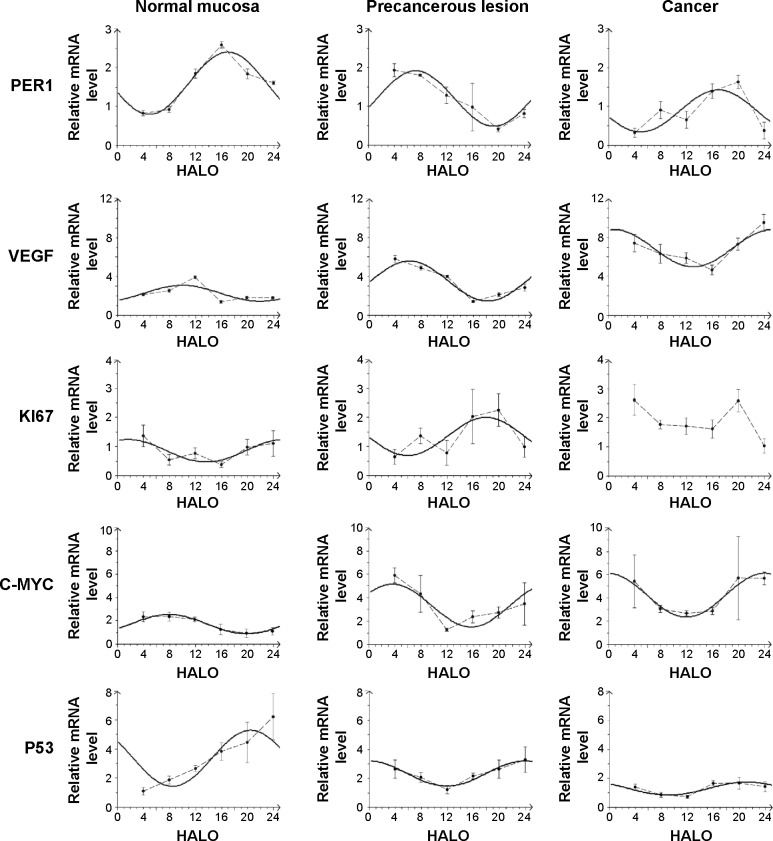
Significant differences in PER1 mRNA expression were noted among the six different time points in the normal, precancerous lesions, and cancer groups (Table 2). The cosine analysis revealed that PER1 exhibited daily rhythmic expression in the normal, precancerous lesions, and cancer groups (Table 2). The cosine-fitted curves are presented in Figure 2.
Table 2.
The mRNA expression of PER1, VEGF, KI67, C-MYC, and P53 in normal buccal mucosa, precancerous lesion, and cancer at six different time points in 24 hours (mean ± SD, n=5)
| Gene | Group | Time point
|
P1-value | P2-value | P3-value | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 4 HALO | 8 HALO | 12 HALO | 16 HALO | 20 HALO | 24 HALO | |||||
| PER1 | N | 0.82±0.07 | 0.91±0.08 | 1.84±0.12 | 2.58±0.08 | 1.83±0.13 | 1.60±0.05 | 0.000 | 0.000 | 0.000a (F=73.324) |
| P | 1.93±0.17 | 1.80±0.03 | 1.28±0.21 | 0.98±0.61 | 0.41±0.07 | 0.81±0.11 | 0.000 | 0.000 | ||
| C | 0.32±0.11 | 0.91±0.22 | 0.66±0.22 | 1.39±0.19 | 1.63±0.17 | 0.38±0.22 | 0.000 | 0.003 | ||
| VEGF | N | 2.12±0.14 | 2.53±0.18 | 3.91±0.16 | 1.39±0.15 | 1.81±0.14 | 1.79±0.12 | 0.000 | 0.0062 | 0.000a (F=469.509) |
| P | 5.81±0.36 | 4.87±0.22 | 3.99±0.12 | 1.42±0.11 | 2.13±0.21 | 2.84±0.36 | 0.000 | 0.000 | ||
| C | 7.43±0.89 | 6.34±0.97 | 5.89±0.54 | 4.67±0.51 | 7.33±0.63 | 9.51±0.84 | 0.000 | 0.0002 | ||
| KI67 | N | 1.37±0.37 | 0.54±0.18 | 0.77±0.19 | 0.37±0.10 | 0.97±0.27 | 1.10±0.44 | 0.010 | 0.0115 | 0.000a (F=31.144) |
| P | 0.64±0.25 | 1.36±0.27 | 0.78±0.43 | 2.03±0.93 | 2.25±0.56 | 0.99±0.36 | 0.011 | 0.0246 | ||
| C | 2.62±0.53 | 1.77±0.15 | 1.71±0.28 | 1.62±0.31 | 2.59±0.39 | 1.03±0.25 | 0.001 | 0.9183 | ||
| C-MYC | N | 2.37±0.43 | 2.38±0.38 | 2.14±0.20 | 1.28±0.45 | 0.96±0.36 | 1.13±0.31 | 0.001 | 0.000 | 0.000b (F=19.542) |
| P | 5.88±0.65 | 4.30±1.59 | 1.72±0.86 | 2.33±0.58 | 2.69±0.68 | 3.45±1.79 | 0.008 | 0.0026 | ||
| C | 5.41±2.29 | 3.04±0.30 | 2.66±0.24 | 2.85±0.29 | 5.70±3.59 | 5.67±0.56 | 0.052 | 0.0128 | ||
| P53 | N | 1.09±0.26 | 1.86±0.27 | 2.64±0.22 | 3.82±0.62 | 4.44±1.40 | 6.20±1.59 | 0.000 | 0.0032 | 0.000a (F=48.358) |
| P | 2.64±0.63 | 2.06±0.33 | 1.23±0.28 | 2.15±0.20 | 2.63±0.62 | 3.28±0.87 | 0.011 | 0.007 | ||
| C | 1.37±0.21 | 0.84±0.17 | 0.71±0.10 | 1.63±0.17 | 1.65±0.42 | 1.40±0.34 | 0.003 | 0.0021 | ||
Notes: P<0.05 represents that there is a significant difference. P1 represents the results of the differences in the expression of each gene at six time points in each group analyzed by one-way ANOVA. P2 represents the results of the daily rhythm of expressions of each gene in each group analyzed by single cosine test. P3 represents the results of the average differences in the expressions of each gene in three groups analyzed by one-way ANOVA. Superscripts “a” and “b” represent the results of the intergroup differences analyzed by Student–Newman–Keuls test after the one-way ANOVA, as follows:
There are mutual significant differences in three groups.
There are significant differences between P or C group and N group, but there is no significant difference between P and C groups.
Abbreviations: SD, standard deviation; n, numbers; HALO, hours after light onset; N, normal buccal mucosa; P, precancerous lesion; C, cancer; ANOVA, analysis of variance.
Figure 2.
Cosine-fitted curves of PER1, VEGF, KI67, C-MYC, and P53 expression in normal buccal mucosa, precancerous lesions, and cancer tissues.
Note: The solid line in the figure represents the cosine-fitted curve, whereas the dotted line connects the mean values of the mRNA expression of each gene at six time points during a 24-hour period.
Abbreviation: HALO, hours after light onset.
The daily rhythms demonstrated the following characteristics (Table 3). 1) Mesor and amplitude: The mesor and amplitude of PER1 mRNA expression in the precancerous lesions group and cancer group were significantly reduced compared with the normal group (P<0.05). The mesor and amplitude of PER1 mRNA in the cancer group were remarkably reduced compared with the precancerous lesions group (P<0.05). 2) Acrophase: The acrophases expressed by PER1 mRNA during various stages of carcinogenesis are presented in Table 3. In the cancer group, the acrophase of PER1 mRNA was approximately identical to that observed in the normal group. In the precancerous lesions group, the acrophase of PER1 mRNA expression was 9.66 hours earlier than that in the normal group.
Table 3.
The daily rhythm characteristics of PER1, VEGF, KI67, C-MYC, and P53 mRNA in normal buccal mucosa, precancerous lesion, and cancer
| Gene | Cosinor parameters | Tissue
|
P-value | ||
|---|---|---|---|---|---|
| N | P | C | |||
| PER1 | Mesor | 1.60±0.11 | 1.20±0.06 | 0.88±0.04 | 0.000a (F=48.137) |
| Amplitude | 0.81±0.01 | 0.71±0.01 | 0.54±0.04 | 0.000a (F=92.659) | |
| Acrophase (HALO) | 16.93±0.04 | 7.27±0.06 | 16.73±0.18 | 0.000c (F=7,294.436) | |
| VEGF | Mesor | 2.26±0.21 | 3.51±0.33 | 6.86±0.59 | 0.000a (F=71.351) |
| Amplitude | 0.82±0.01 | 2.06±0.23 | 1.91±0.06 | 0.000b (F=71.938) | |
| Acrophase (HALO) | 9.93±0.91 | 6.21±0.24 | 1.04±0.95 | 0.000a (F=100.324) | |
| KI67 | Mesor | 0.85±0.42 | 1.34±0.76 | – | 0.000a (F=12.387) |
| Amplitude | 0.39±0.07 | 0.66±0.03 | – | 0.013a (F=17.970) | |
| Acrophase (HALO) | 1.65±0.71 | 19.93±0.74 | – | 0.003a (F=42.501) | |
| C-MYC | Mesor | 1.71±0.10 | 3.39±0.03 | 4.22±0.11 | 0.000a (F=543.026) |
| Amplitude | 0.83±0.06 | 1.74±0.01 | 1.87±0.04 | 0.000b (F=574.424) | |
| Acrophase (HALO) | 7.78±0.57 | 3.92±0.55 | 23.93±0.06 | 0.000a (F=1,607.264) | |
| P53 | Mesor | 3.34±0.19 | 2.33±0.26 | 1.27±0.21 | 0.000a (F=82.040) |
| Amplitude | 1.92±0.13 | 0.86±0.08 | 0.45±0.07 | 0.000a (F=109.770) | |
| Acrophase (HALO) | 20.47±0.81 | 23.87±0.89 | 21.06±0.73 | 0.005c (F=14.995) | |
Notes: The daily rhythm is described as mesor, amplitude, and acrophase. P<0.05 represents that there is a significant difference. P-value represents the results of differences in each cosinor parameter of each gene in three groups analyzed by one-way ANOVA. Superscripts “a”, “b”, and “c” represent the results of the intergroup differences analyzed by Student–Newman–Keuls test after the one-way ANOVA, as follows:
There are mutual significant differences in three groups.
There are significant differences between P or C group and N group, but there is no significant difference between P and C groups.
There are significant differences between P group and C or N group, but there is no significant difference between C and N groups.
Abbreviations: N, normal buccal mucosa; P, precancerous lesion; C, cancer; HALO, hours after light onset; ANOVA, analysis of variance.
Daily rhythm variations of mRNA of cancer-related genes during different stages of buccal mucosa carcinogenesis
Significant differences in VEGF, KI67, and P53 mRNA were noted among the six daily time points in the normal, precancerous lesions, and cancer groups, respectively (P<0.05). C-MYC mRNA exhibited significant differences at the six daily time points in the normal and precancerous lesions groups (P<0.05); however, no differences were noted in the cancer group (P>0.05), as shown in Table 2. The cosine analysis indicated that VEGF, P53, and C-MYC mRNA, respectively, exhibited daily rhythmic expression in the normal, precancerous lesions, and cancer groups (P<0.05). KI67 mRNA exhibited daily rhythmic expression in the normal and precancerous lesions groups (P<0.05), whereas this expression pattern was not observed in the cancer group (P>0.05) (Table 2 and Figure 2).
The daily rhythms exhibited the following characteristics (Table 3). 1) Mesor: The mesor of VEGF and C-MYC mRNA expression in the precancerous lesions and cancer groups, respectively, was significantly increased compared with the normal group (P<0.05). In the cancer group, the mesor of VEGF and C-MYC mRNA was remarkably increased compared with that in the precancerous lesions group (P<0.05). The mesor of KI67 mRNA in the precancerous lesions group was remarkably increased compared with the normal group (P<0.05). The mesor of P53 mRNA in the precancerous lesions group and cancer group significantly decreased compared with the normal group (P<0.05). In the cancer group, the mesor was significantly reduced compared with the precancerous lesions group (P<0.05). 2) Amplitude: The amplitudes of VEGF and C-MYC mRNA in the precancerous lesions and cancer groups, respectively, were significantly increased compared with the normal group (P<0.05). No remarkable difference was noted between the precancerous lesions and cancer groups (P>0.05). The amplitude of KI67 mRNA in the precancerous lesions group was remarkably increased compared with the normal group (P<0.05). The amplitude of P53 mRNA in the precancerous lesions and cancer groups was significantly reduced compared with the normal group (P<0.05), and the amplitude in the cancer group was remarkably decreased compared with the precancerous lesions group (P<0.05). 3) Acrophase: The acrophases of VEGF, KI67, C-MYC, and P53 mRNA in various carcinogenesis periods are presented in Table 3. The acrophases of VEGF mRNA in the precancerous lesions and cancer groups were 3.72 hours and 8.89 hours earlier, respectively, compared with the normal group. The acrophase of KI67 mRNA in the precancerous lesions group was delayed 18.28 hours compared with the normal group. The acrophases of P53 mRNA were similar in the normal and cancer groups. In the precancerous lesions group, the acrophase was 3.4 hours later than that observed in the normal group. The acrophases of C-MYC mRNA in the precancerous lesions and cancer groups were 3.86 hours earlier and 16.15 hours later, respectively, compared with the normal group.
Discussion
Current studies have indicated that the clock gene PER1 is a cancer suppressor gene, and its circadian variations exhibit a close relationship with the occurrence and development of cancers.8,15,27–29,32 Bjarnason et al reported that PER1 expression in human buccal mucosa cells exhibits a circadian rhythm.25 Our previous research demonstrated decreased and circadian rhythmic PER1 expression in oral squamous cell carcinomas.28,33 This study reports that the expression of PER1 in three stages of oral squamous cell carcinoma exhibited daily rhythms. The mesor and amplitude of PER1 mRNA expression decreased remarkably as carcinogenesis progressed. These results demonstrate that the role of PER1 in cancer inhibition was gradually reduced, probably resulting in malignant transformation. In terms of the acrophase, the timing of PER1 mRNA expression peak values was similar in the normal buccal mucosa and carcinoma stages, and the patterns of PER1 mRNA daily rhythmic expression were similar. In the precancerous lesions stage, the acrophase of PER1 mRNA was 9.66 hours earlier than that observed in the normal buccal mucosa. The expression pattern of the daily rhythms of PER1 mRNA in the normal buccal mucosa was in contrast to that observed in precancerous lesions. The mechanism of these variations requires further study.
Carcinogenesis is dependent on tumor angiogenesis, and VEGF plays a central role in tumor-induced angiogenesis.34,35 Koyanagi et al reported that VEGF mRNA expression in transplanted sarcoma cells of mice exhibits circadian rhythms. In hepatic cells and plasma of mice with tumors, VEGF protein exhibits circadian rhythmic expression.36 Our research found that VEGF mRNA expression exhibits daily rhythms in the three carcinogenesis stages in oral squamous cell carcinoma. The mesor and amplitude of VEGF mRNA expression were significantly elevated upon the development of cancer, thus promoting tumor angiogenesis and carcinogenesis. The acrophase of VEGF mRNA constantly progressed with the occurrence and development of cancer. The mechanism of such variations in carcinogenesis requires further exploration and serves as an important reference value for the administration of cancer chemotherapies using VEGF as a biomarker.
P53 is a cancer suppressor gene, and increased P53 expression inhibits tumor growth and induces cell apoptosis.37,38 KI67 and C-MYC are cell proliferation genes that promote cell proliferation via increases in their expression.37,39 Our research demonstrated that P53 and C-MYC mRNA exhibit daily rhythmic expression in three stages of buccal mucosa carcinogenesis. However, KI67 mRNA exhibited daily rhythmic expression in the normal and precancerous lesions groups, whereas KI67 mRNA expression was dysregulated in the cancer group. During the occurrence and development of cancer, the mesor and amplitude of P53 mRNA expression typically decreased gradually, indicating a weakened effect on cancer inhibition and apoptotic induction. KI67 and C-MYC mRNA expression increased gradually, resulting in enhanced reinforcement of cell proliferation and leading to an imbalance between cell proliferation and apoptosis. Regarding the acrophase, P53 mRNA expression was approximately identical in the normal and cancer groups; P53 mRNA expression in the precancerous lesions group lagged by 3.4 hours compared with the normal group. In contrast, the acrophase of KI67 mRNA in the precancerous lesions group was 18.28 hours later than the normal group. C-MYC mRNA expression was 3.86 hours earlier and 16.15 hours later in the precancerous lesions and cancer groups, respectively. The uncoordinated changes in the acrophases of the apoptosis gene P53 and cell proliferation genes KI67 and C-MYC probably further aggravate the imbalance between cell proliferation and apoptosis, thus promoting carcinogenesis.
Conclusion
The specific mechanisms by which the clock gene PER1 regulates and controls cancer-related genes, such as VEGF, KI67, C-MYC, and P53, and the characteristic changes in the circadian rhythms of these genes remain unclear; however, this study was the first to demonstrate the daily rhythmic expression patterns of the clock gene PER1 and cancer-related genes, such as VEGF, KI67, C-MYC, and P53, in different stages of carcinogenesis. These results indicate that the mesors, amplitudes, and acrophases of the clock gene PER1 and cancer-related genes VEGF, KI67, C-MYC, and P53 undergo substantial changes upon the development of cancer. This study provides novel ideas and methods for the exploration of carcinogenic mechanisms and cancer treatments from the perspective of the relationship between circadian variations in gene expression and carcinogenesis.
Acknowledgments
We thank Miss Ling Li for her help in preparing the manuscript and Mr Baisong Li for assisting in the statistical analysis of the data.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Zieker D, Jenne I, Koenigsrainer I, et al. Circadian expression of clock- and tumor suppressor genes in human oral mucosa. Cell Physiol Biochem. 2010;26(2):155–166. doi: 10.1159/000320547. [DOI] [PubMed] [Google Scholar]
- 2.Borgs L, Beukelaers P, Vandenbosch R, Belachew S, Nguyen L, Malgrange B. Cell “circadian” cycle: new role for mammalian core clock genes. Cell Cycle. 2009;8(6):832–837. doi: 10.4161/cc.8.6.7869. [DOI] [PubMed] [Google Scholar]
- 3.Bozek K, Relógio A, Kielbasa SM, et al. Regulation of clock-controlled genes in mammals. PLoS One. 2009;4(3):e4882. doi: 10.1371/journal.pone.0004882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wulff K, Porcheret K, Cussans E, Foster RG. Sleep and circadian rhythm disturbances: multiple genes and multiple phenotypes. Curr Opin Genet Dev. 2009;19(3):237–246. doi: 10.1016/j.gde.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Oster H, Damerow S, Hut RA, Eichele G. Transcriptional profiling in the adrenal gland reveals circadian regulation of hormone biosynthesis genes and nucleosome assembly genes. J Biol Rhythms. 2006;21(5):350–361. doi: 10.1177/0748730406293053. [DOI] [PubMed] [Google Scholar]
- 6.Keller M, Mazuch J, Abraham U, et al. A circadian clock in macrophages controls inflammatory immune responses. Proc Natl Acad Sci USA. 2009;106(50):21407–21412. doi: 10.1073/pnas.0906361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang XL, Fu CJ, Bu RF. Role of circadian clocks in the development and therapeutics of cancer. J Int Med Res. 2011;39(6):2061–2066. doi: 10.1177/147323001103900601. [DOI] [PubMed] [Google Scholar]
- 8.Cao Q, Gery S, Dashti A, et al. A role for the clock gene per1 in prostate cancer. Cancer Res. 2009;69(19):7619–7625. doi: 10.1158/0008-5472.CAN-08-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bjarnason GA, Jordan RC, Sothern RB. Circadian variation in the expression of cell-cycle proteins in human oral epithelium. Am J Pathol. 1999;154(2):613–622. doi: 10.1016/S0002-9440(10)65306-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rana S, Mahmood S. Circadian rhythm and its role in malignancy. J Circadian Rhythms. 2010;8:3. doi: 10.1186/1740-3391-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Savvidis C, Koutsilieris M. Circadian rhythm disruption in cancer biology. Mol Med. 2012;18:1249–1260. doi: 10.2119/molmed.2012.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111(7):919–922. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- 13.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 14.Schibler U, Ripperger J, Brown SA. Peripheral circadian oscillators in mammals: time and food. J Biol Rhythms. 2003;18(3):250–260. doi: 10.1177/0748730403018003007. [DOI] [PubMed] [Google Scholar]
- 15.Soták M, Polidarová L, Ergang P, Sumová A, Pácha J. An association between clock genes and clock-controlled cell cycle genes in murine colorectal tumors. Int J Cancer. 2013;132(5):1032–1041. doi: 10.1002/ijc.27760. [DOI] [PubMed] [Google Scholar]
- 16.Yoo SH, Yamazaki S, Lowrey PL, et al. PERIOD2: luciferase real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101(15):5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shearman LP, Sriram S, Weaver DR, et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288(5468):1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 18.Gekakis N, Staknis D, Nguyen HB, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1988;280(5369):1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 19.Kume K, Zylka MJ, Sriram S, et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98(2):193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 20.Sato TK, Yamada RG, Ukai H, et al. Feedback repression is required for mammalian circadian clock function. Nat Genet. 2006;38(3):312–319. doi: 10.1038/ng1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Storch KF, Lipan O, Leykin I, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417(6884):78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 22.Meyer V, Lerchl A. Evidence for species-specific clock gene expression patterns in hamster peripheral tissues. Gene. 2014;548(1):101–111. doi: 10.1016/j.gene.2014.07.019. [DOI] [PubMed] [Google Scholar]
- 23.Rana S, Munawar M, Shahid A, et al. Deregulated expression of circadian clock and clock-controlled cell cycle genes in chronic lymphocytic leukemia. Mol Biol Rep. 2014;41(1):95–103. doi: 10.1007/s11033-013-2841-7. [DOI] [PubMed] [Google Scholar]
- 24.Fu L, Kettner NM. The circadian clock in cancer development and therapy. Prog Mol Biol Transl Sci. 2013;119:221–282. doi: 10.1016/B978-0-12-396971-2.00009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bjarnason GA, Jordan RC, Wood PA, et al. Circadian expression of clock genes in human oral mucosa and skin: association with specific cell-cycle phases. Am J Pathol. 2001;158(5):1793–1801. doi: 10.1016/S0002-9440(10)64135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng L, Seon YJ, McHugh J, Papagerakis S, Papagerakis P. Clock genes show circadian rhythms in salivary glands. J Dent Res. 2012;91(8):783–788. doi: 10.1177/0022034512451450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lengyel Z, Lovig C, Kommedal S, et al. Altered expression patterns of clock gene mRNAs and clock proteins in human skin tumors. Tumor Biol. 2013;34(2):811–819. doi: 10.1007/s13277-012-0611-0. [DOI] [PubMed] [Google Scholar]
- 28.Zhao N, Yang K, Yang G, et al. Aberrant expression of clock gene period1 and its correlations with the growth, proliferation and metastasis of buccal squamous cell carcinoma. PLoS One. 2013;8(2):e55894. doi: 10.1371/journal.pone.0055894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell. 2006;22(3):375–382. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 30.Pluquet O, Dejeans N, Chevet E. Watching the clock: endoplasmic reticulum-mediated control of circadian rhythms in cancer. Ann Med. 2014;46(4):233–243. doi: 10.3109/07853890.2013.874664. [DOI] [PubMed] [Google Scholar]
- 31.WHO Collaborating Centre for Oral Precancerous Lesions Definition of leukoplakia and related lesions: an aid to studies on oral precancer. Oral Surg Oral Med Oral Pathol. 1978;46:518–539. [PubMed] [Google Scholar]
- 32.Lee S, Donehower LA, Herron AJ, Moore DD, Fu L. Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. PLoS One. 2010;5(6):e10995. doi: 10.1371/journal.pone.0010995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen R, Yang K, Zhao NB, et al. Abnormal expression of PER1 circadian-clock gene in oral squamous cell carcinoma. Onco Targets Ther. 2012;5:403–407. doi: 10.2147/OTT.S38508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang CY, Wen MS, Wang HW, et al. Increased vascular senescence and impaired endothelial progenitor cell function mediated by mutation of circadian gene Per2. Circulation. 2008;118(21):2166–2173. doi: 10.1161/CIRCULATIONAHA.108.790469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood PA, Du-Quiton J, You S, Hrushesky WJ. Circadian clock coordinates cancer cell cycle progression, thymidylate synthase, and 5-fluorouracil therapeutic index. Mol Cancer Ther. 2006;5(8):2023–2033. doi: 10.1158/1535-7163.MCT-06-0177. [DOI] [PubMed] [Google Scholar]
- 36.Koyanagi S, Kuramoto Y, Nakagawa H, et al. A molecular mechanism regulating circadian expression of vascular endothelial growth factor in tumor cells. Cancer Res. 2003;63(21):7277–7283. [PubMed] [Google Scholar]
- 37.Granda TG, Liu XH, Smaaland R, et al. Circadian regulation of cell cycle and apoptosis proteins in mouse bone marrow and tumor. FASEB J. 2005;19(2):304–306. doi: 10.1096/fj.04-2665fje. [DOI] [PubMed] [Google Scholar]
- 38.Rengarajan T, Nandakumar N, Rajendran P, Haribabu L, Nishigaki I, Balasubramanian MP. D-pinitol promotes apoptosis in MCF-7 cells via induction of p53 and Bax and inhibition of Bcl-2 and NF-κB. Asian Pac J Cancer Prev. 2014;15(4):1757–1762. doi: 10.7314/apjcp.2014.15.4.1757. [DOI] [PubMed] [Google Scholar]
- 39.Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182(3):311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]