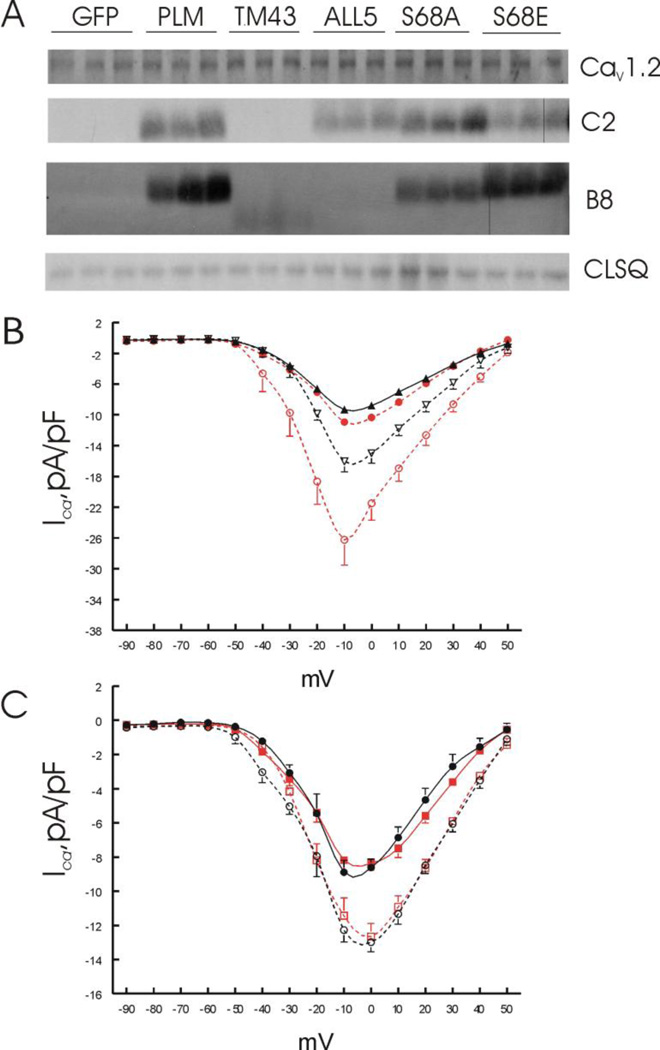
Figure 3.
Effects of PLM mutants on Cav1.2 expression and ICa. LV myocytes isolated from KO hearts were infected with adenovirus (Adv) expressing either green fluorescent protein (GFP), WT PLM, the cytoplasmic domain truncation mutant TM43, the signature PFXYD motif changed to Ala mutant (ALL5), the non-phosphorylable Ser68 mutant (S68A), or the phosphomimetic Ser68 mutant (S68E); and placed in culture for 24 hours prior to ICa measurements and Western blotting. A. Myocyte lysates were prepared and probed for Cav1.2, C-terminus (C2) or N-terminus (B8) of PLM and its mutants, with calsequestrin (CLSQ) serving as the loading control. There were no differences (p<0.98) in Cav1.2 expression among KO myocytes expressing GFP (1.44 ± 0.01), WT PLM (1.42 ±0.01), TM43 (1.45 ± 0.08), ALL5 (1.46 ± 0.07), S68A (1.42 ± 0.00) and S68E (1.46 ± 0.05) mutants (n=3 each; values in arbitrary units). As expected, C2 which detects the cytoplasmic tail of PLM fail to detect any signal in KO-GFP and KO-TM43 myocytes. Likewise, there were no B8 (detects the extracellular NH2-terminus of dog PLM) signals in KO-GFP and KO-ALL5 myocytes. B8 signals in KO-TM43 myocytes were weak, downward shifted but present. B. I-V curves of ICa in KO-GFP (●; n=8) and KO-PLM myocytes (▲; n=6) at baseline; and after exposure to 1 µM isoproterenol in KO-GFP (○; n=6) and KO-PLM (∇; n=4) myocytes. C. I-V curves of ICa in KO-S68A (●; n=6) and KO-S68E (■; n=6) myocytes at baseline, and after addition of 1 µM isoproterenol to KO-S68A (○; n=5) and KO-S68E (□; n=5) myocytes. Note the difference in ordinate scales between B and C.