FIGURE 7.
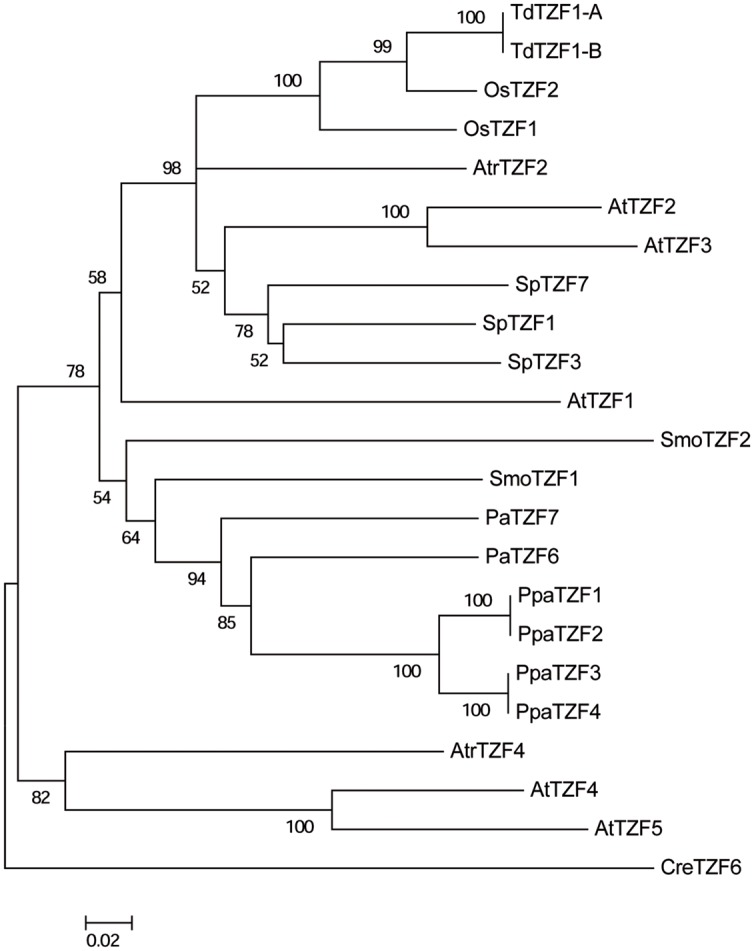
Phylogenetic tree of the AtTZF1-2-3-4-5-like proteins from selected species. The unrooted tree was constructed using the neighbor-joining method after alignment of the full-length amino-acid sequences using the Clustal W algorithm. Bootstrap values from 1,000 replicates are indicated at each node, and only bootstrap values higher than 50% from 1,000 replicates are shown. Scale bar: estimated 0.2 amino-acid substitutions per site.
