Figure 5.
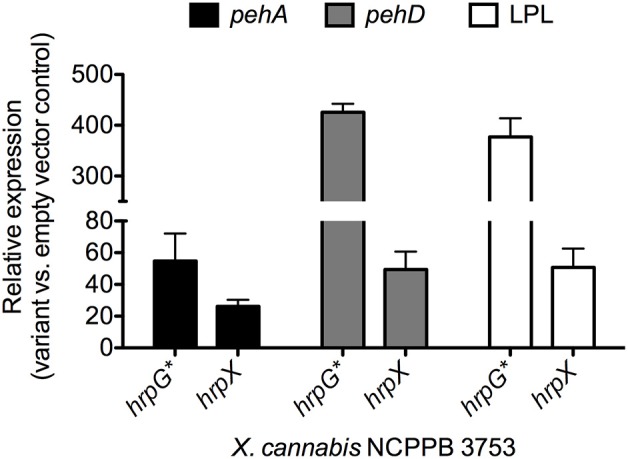
Gene expression analysis of genes with HrpX-inducible PIP boxes. Bacteria were grown overnight in liquid NB (Sigma Aldrich, USA) supplemented with gentamycin (20 μg l−1) and transferred to fresh 10 mL NB media with gentamycin for a final OD600 = 0.5. Bacteria were incubated for 3 h, shaking at 28°C. Transcriptional profiles and RNA was preserved with 5% phenol in ethanol as previously described (Jahn et al., 2008; Jacobs et al., 2012). Bacterial RNA was extracted with Trizol (Invitrogen, USA), cleaned up with Zymogen RNA concentrator (Zymo Research, USA) and treated with Turbo DNase (Invitrogen, USA) following manufacturer's protocols. RNA (1 μg per sample) was reverse transcribed with Superscript III (Invitrogen, USA) following the manufacturer's recommendation. qPCR with SYBR MESA BLUE Master Mix (Eurogentec, Belgium) was performed following the manufacturer's protocol on a Roche LightCycler 480 Real-Time PCR instrument (Roche Diagnostics Corporation, USA) with reaction parameters of 10-min polymerase activation at 95°C, then 40 cycles, with an individual cycle consisting of 15 s at 95°C and 1 min at 60°C. Relative gene expression analysis of pehA, pehD and the gene encoding the putative lysophospholipase (LPL) were calculated by the delta-delta-Ct method (Livak and Schmittgen, 2001) for NCPPB 3753 variants ectopically expressing hrpG* or hrpX compared to NCPPB 3753 pBBR1MCS-5. Expression of atpD was used as a normalization internal control gene. Primers are listed in Supplemental Table S1.
