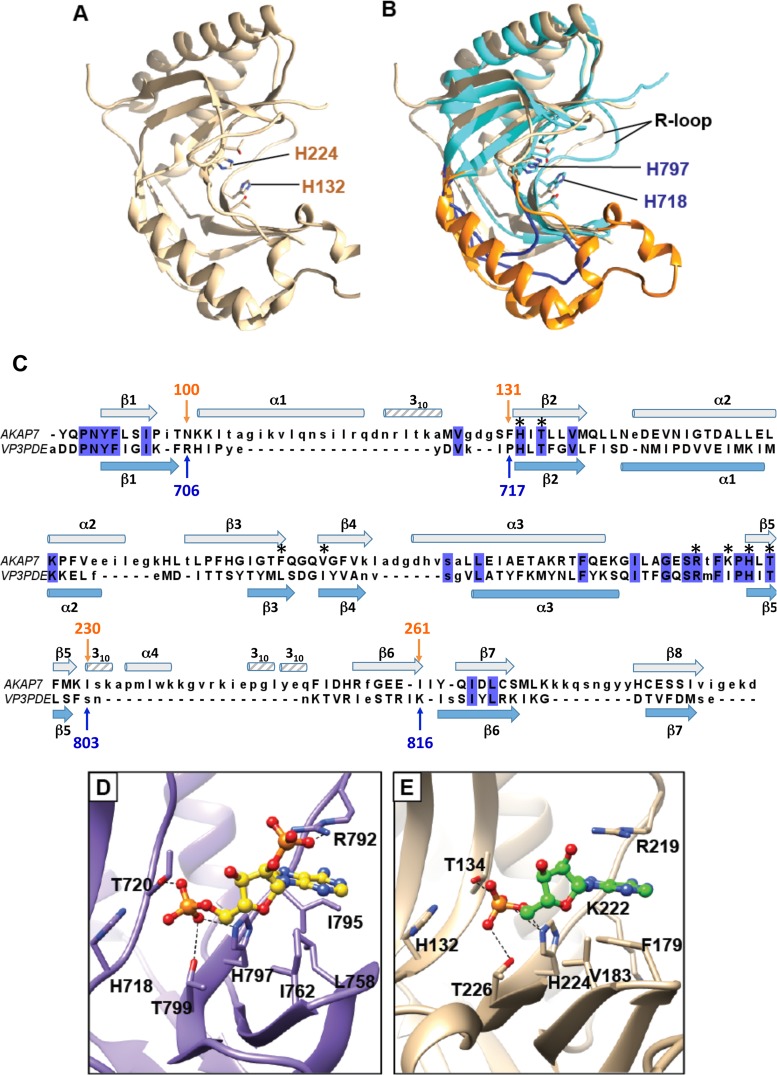
FIG 5.
Structural homolog of the RVA VP3 CTD. AKAP7 (tan; PDB 2VFK) is shown alone (A) or superimposed on the RVA VP3 CTD (cyan) (B). Two loops of the RVA VP3 CTD, residues 706 to 717 and 803 to 816, are shown in dark blue. The corresponding AKAP7 residues, 100 to 131 and 230 to 261, which form helices and strands, are shown in orange. The R loop in each structure is indicated. (C) Structure-based alignment of RVA VP3 CTD and AKAP7 central domain. Blue highlighting indicates identical amino acids in the alignment. The secondary structure of the RVA VP3 CTD is shown in light blue below the sequences, and the secondary structure of the AKAP7 central domain is shown in gray above. Asterisks denote VP3 CTD residues that interact with 2-5A (Fig. 4). (D and E) Comparison of the catalytic sites of the RVA VP3 CTD (D; purple) and AKAP7 (E; tan). RVA VP3 CTD residues interacting with 2-5A (yellow ball-and-stick representation) are labeled. AKAP7 residues interacting with AMP (green ball-and-stick representation) are shown. Blue atoms, N; red atoms, O; orange atoms, P.