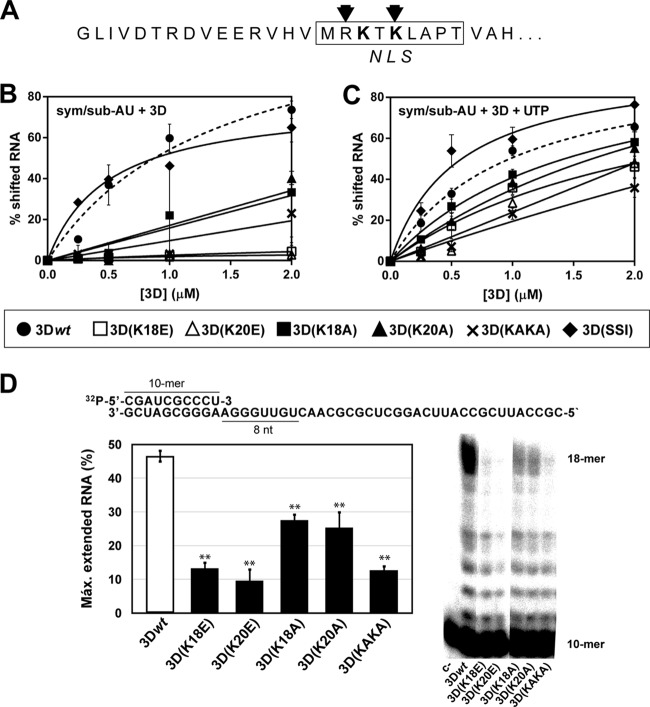
FIG 1.
RNA binding and processivity of wild-type and mutant 3Ds. (A) Amino acid sequence of the 27 amino-terminal residues of 3D. The box delimits the sequence identified as a nuclear localization signal (NLS) (13), the arrows indicate the amino acids that interact with RNA (12), and boldface highlights the substituted residues. (B) Electrophoretic mobility shift assay without UTP, where the indicated concentrations of 3D proteins were incubated with labeled sym/sub-AU (CGUAGGGCCC) RNA, as described in Materials and Methods. The products were separated electrophoretically in a 5% polyacrylamide gel; the percentage of shifted labeled RNA was plotted as a function of protein concentration, and the data points were fit to one-site binding (hyperbola). Binding of 3Dwt to RNA is represented as a dashed line. The values are the average of three determinations, and standard deviations are given. (C) Electrophoretic mobility shift assay with UTP. Reactions were performed as described in panel B except for the presence of 50 μM UTP during the incubation period. (D) The sequence of the heteropolymeric template (10-mer)-primer used in the processivity assay is given at the top; the sequence copied by 3D in the presence of ATP, UTP and CTP (absence of GTP) is underlined. The graph shows a representative electrophoretic separation (23% denaturing PAGE) of the products of elongation after a 5-min reaction with ATP, CTP, and UTP. The values represent the percentages of extended 18-mers measured relative to the total amount of elongated RNA (all bands shown in the electropherograms except the 10-mer). The values are the averages of three determinations, and standard deviations are given. Asterisks indicate the statistical significance of the difference with the value for 3Dwt (**, P < 0.0001; analysis of variance test).