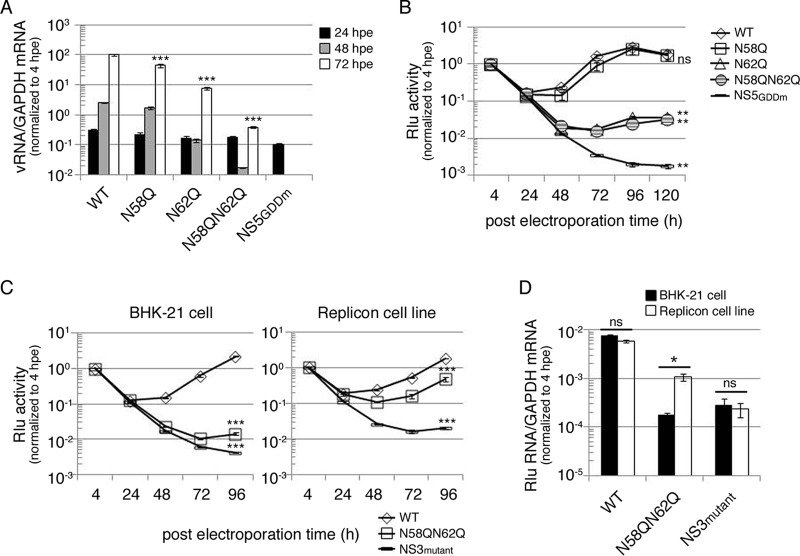
FIG 5.
Mutation of putative N-glycosylation sites on DENV NS4B decreased viral RNA replication. (A) Replication analysis of NS4B mutants using full-length (FL) infectious clone RNA. FL RNA (10 μg) encoding WT, N58Q, N62Q, or N58QN62Q NS4B was electroporated into BHK-21 cells (3 × 106 cells). The NS5GDDm FL RNA was a replication-deficient control. At the indicated time points postelectroporation, total cellular RNA was isolated. The viral RNA NS3 protein coding region was quantitated using real-time qPCR, and host GAPDH mRNA quantification served as a control. All the later time points were normalized with the 4-hpe value. Asterisks (***, P < 0.0005) indicate results significantly different from the WT. The data for the last time point in the experiment were chosen for statistical analysis. (B) Replication analysis of NS4B mutants using DENV subgenomic replicon with Renilla luciferase reporter. DENV SGR-Rlu replicon RNA (10 μg) encoding WT, N58Q, N62Q, or N58QN62Q NS4B was electroporated into BHK-21 cells (3 × 106 cells). The NS5GDDm SGR-Rlu replicon was a replication-deficient control. Cell lysate was prepared at 4, 24, 48, 72, 96, and 120 hpe to determine the Renilla luciferase activity at each time point. All the later time points were normalized against 4-h Renilla luciferase activity that indicated the electroporation efficiency. The normalized Renilla luciferase activity at different time points is expressed as the mean ± standard deviation of triplicates. The experiment was performed in triplicate, and the data shown are from one representative electroporation experiment. Asterisks (**, P < 0.005) indicate results significantly different from the WT, and “ns” represents no significant difference. The data for the last time point in the experiment were chosen for statistical analysis. (C and D) trans-complementation analyses of NS4B N58QN62Q mutant by Renilla luciferase reporter (C) and real-time qPCR (D). For panel C, naive BHK-21 cells or the D2-Flu-SGR-Neo replicon cell line was electroporated with DENV SGR-Rlu replicon RNA (10 μg) encoding WT NS4B, N58QN62Q mutant NS4B, or NS3 K199AT200A mutant. The normalized Renilla luciferase activity at different time points postelectroporation is expressed as mean ± standard deviation for triplicate experiments. The experiment was performed in triplicate, and the data shown are from one representative experiment. Asterisks (***, P < 0.0005) indicate results significantly different from the WT. The data from the last time point in the experiment were chosen for statistical analysis. For panel D, at 4 hpe and 96 hpe, total RNA was isolated from both host cells. The Renilla luciferase region was amplified with qPCR primers, GAPDH mRNA quantification served as a control, and 96-hpe data were normalized to the 4-hpe value. An asterisk (*, P < 0.05) indicates a significant difference from the naive BHK-21 cells electroporated with DENV SGR-Rlu replicon NS4B N58QN62Q mutant RNA, and “ns” represents no significant difference. The experiment was performed twice, and the data shown are from one representative electroporation experiment.