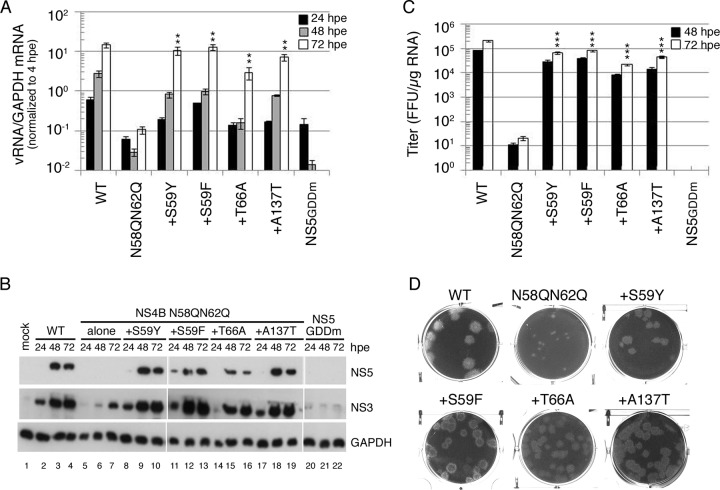
FIG 7.
Rescuing effect of compensatory mutations on NS4B N58QN62Q mutation. (A) Viral RNA synthesis. BHK-21 cells were electroporated with FL RNA (10 μg) encoding WT, N58QN62Q, and N58QN62Q+suppressor NS4B. The levels of viral RNA and cellular GAPDH mRNA at 4, 24, 48, and 72 hpe were quantified by real-time qPCR. The data were normalized to the 4-hpe value, which reflects the FL RNA electroporation efficiency. The electroporation experiment was performed three times, and real-time qPCR was done in triplicate. The mean ± standard deviation shown here is from one representative electroporation experiment. Asterisks (**, P < 0.005) indicate results significantly different from the NS4B N58QN62Q mutation. The data from the last time point in the experiment were chosen for statistical analysis. (B) Replication-dependent translation of FL RNA. At 24, 48, and 72 hpe, the cells were lysed, and NS3 and NS5 expression was analyzed by Western blotting. GAPDH expression served as a protein loading control. (C) Virus production. Culture fluid of FL RNA electroporated BHK-21 cells collected at 48 hpe and 72 hpe was used to infect naive BHK-21 cells, and virus titer was determined by focus-forming assay. Results are expressed as mean ± standard deviation of triplicates. The experiment was performed three times, and the data shown are from one representative electroporation experiment. Asterisks (***, P < 0.0005) indicate results significantly different from the NS4B N58QN62Q mutation. The data from the last time point in the experiment were chosen for statistical analysis. (D) Plaque morphology. Culture fluid from 144 hpe was subjected to plaque assay in BHK-21 cells. Plaques were fixed at 12 dpi.