Fig. 5.
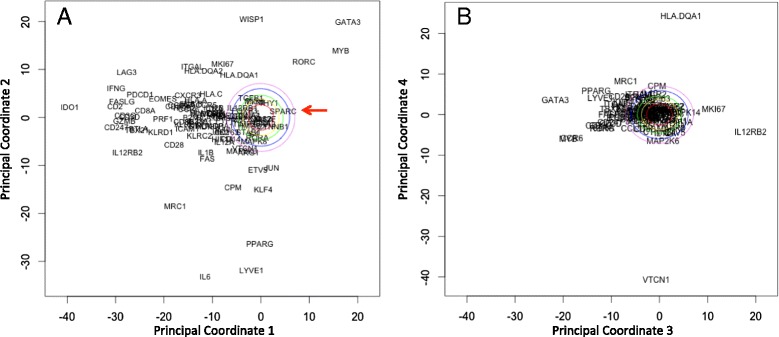
Increased WISP1 expression correlates with increased GATA3 expression. Principal coordinate analysis was applied to expression data for a subset of immune-related genes obtained from the invasive breast cancer arm of the Cancer Genome Atlas (see Figure S3 in [108]). Projections of the genes along the first two principal coordinate (PC) directions (a) and the third and fourth PC directions (b). PC 1 can be interpreted as a type 1 immune signature and PC 2 is interpreted as a signature of oncogenic transformation in invasive breast cancer. The first four principal coordinates capture 20, 13, 7, and 6 % of the overall variance in the data, respectively. As principal coordinates are independent, the projection of a gene along the corresponding axes indicates the degree to which the expression of two genes are related and the distance from the origin indicates the strength of the covariation within the data set. The remaining principal coordinates capture progressively less variance in the data and provide little additional information, as illustrated by the distribution of the genes along PC 4, except HLA.DQA1 and VTCN1, are close to the origin. The colored ovals radiating out from the origin indicate principal coordinate values that can not be distinguished from random noise, that is a null hypothesis, with increasing levels of statistical stringency. The red arrow in panel A indicates SPARC. These colored ovals were obtained using a simulation approach called bootstrap resampling
