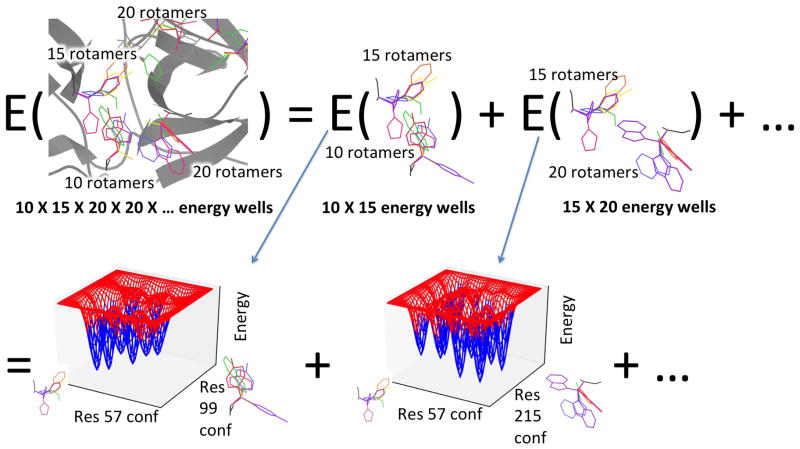
Figure 2.
The number of energy wells in a protein system scales exponentially with the number of flexible residues, leading to an exponential number of energy function calls, but EPIC can replace most of these calls with quick evaluations of low-degree polynomials. (Top) A protein may have an energy well for every combination of rotamers (rainbow) at different residues. The global minimum-energy conformation (GMEC) of a protein may be in any of these wells. We model the energy as a sum of pairwise energy terms. Each pairwise term will have wells for pairs of rotamers, but there are far fewer wells of this kind—a number quadratic in the number of residues. We can easily afford the energy function calls needed to characterize each pairwise well. (Bottom) By precomputing a polynomial representation (blue) of the energy within each well of each pairwise term (red), we enable computation of any pairwise term in any pairwise well, and thus of the full protein energy in any energy well of the protein, solely by a quick evaluation of polynomials.