Figure 5.
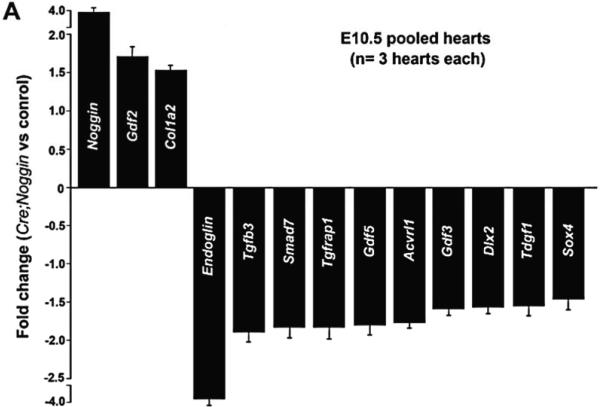
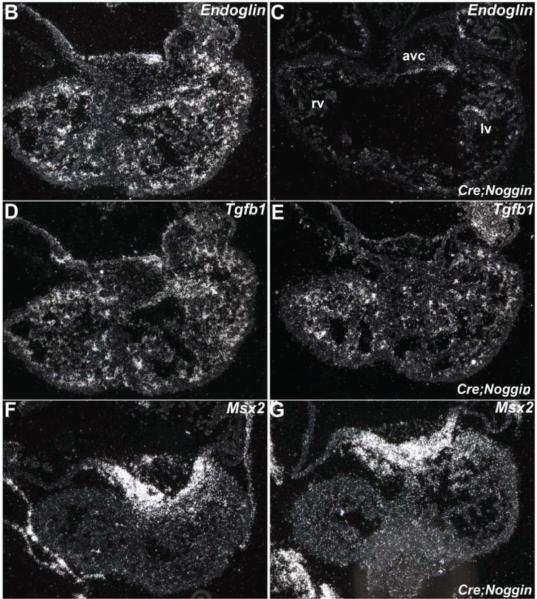
qPCR mRNA profiling and in situ verification of expression alterations. (A): Quantitative PCR using a custom array of 84 genes responsive to TGFß superfamily signal transduction revealed statistically significant changes in transcript levels of the 13 listed genes. Data are presented as a logarithmic plot of relative expression (mutant/wildtype), where a value of 1 indicates no difference between E10.5 NogEnd mutant and wildtype pooled hearts and values <1 indicate reduced expression. The biggest alterations were observed in Noggin levels, which were x4 fold elevated in mutant hearts; and Endoglin levels, which were×3.7 fold reduced in mutant hearts. Error bars represent SEM; (B,C): Reduced Endoglin expression levels were verified by radioactive in situ hybridization, as the E10.5 mutant heart (B) exhibits significantly less Endoglin mRNA throughout the endocardial lineage when compared to wildtype littermate (B); (D–G): As qPCR profiling did not reveal any alterations in either endocardially-restricted Tgfβ1 or myocardial Msx2 expression levels, we used in situ hybridization to examine whether spatial alterations were present in mutant hearts; Radioactive in situ hybridization revealed comparable Tgfβ1 mRNA expression in control (D) and mutants (E), and Msx2 was normally expressed within the controls (F) and NogEnd mutant (F) myocardium adjacent to the AV cushions. Abbreviations: avc, atrioventricular cushion; lv, left ventricle; rv, right ventricle.