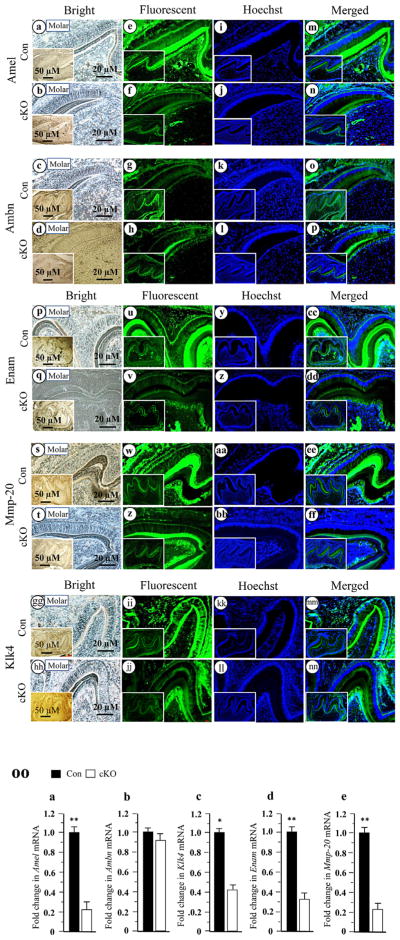
Fig. 6.
Altered expression of mouse enamel matrix and enamel-processing protein genes in the Bmp2 cKO teeth. Tissue sections at PN6 were photographed under a light microscope using a digital cooled camera (a–d, q–t, gg–hh). The expression of enamel matrix and enamel processing proteins were analyzed and quantified by fluorescent immunohistochemistry assay with antibodies specific to Amel, Ambn, Enam, Mmp-20 and Klk4 (e–h, u–x, ii–jj). The tissue sections were stained with Hoechst for the nucleus (i–l, y–bb, kk–ll). Images were merged (m–p, cc–ff, mm–nn). Lower magnifications are shown in insets. Protein expression of Amel, Enam, Mmp-20 and Klk4 were reduced in the Bmp2 cKO enamels, whereas Ambn expression was unaffected. OO. Total RNAs were isolated from the wild-type and Bmp2 cKO teeth at PN6. The mRNA levels of Amel, Ambn, Enam, Mmp-20 and Klk4 were measured by qRT-PCR. Cyclophilin A was used as an internal control. Expression of these mRNAs in the teeth of the control mice acts as a 1.0-fold increase. The bar graphs show mean ± S.D. (n = 3) from three independent experiments. qRT-PCR shows a significant decrease of Amel (a), Klk4 (c), Enam (d) and Mmp-20 (e) mRNAs and no difference in Ambn (b) mRNA expression at PN6 in the Bmp2 cKO mice. Asterisks indicate significant differences between the control and Bmp2 cKO groups (*P <0.05, **P <0.01). Con, control; cKO, Bmp2 cKO.