Abstract
Background
Potentially modifiable risk factors including obesity, diabetes, hypertension, and smoking are associated with Alzheimer disease (AD) and represent promising targets for intervention. However, the causality of these associations is unclear. We sought to assess the causal nature of these associations using Mendelian randomization (MR).
Methods and Findings
We used SNPs associated with each risk factor as instrumental variables in MR analyses. We considered type 2 diabetes (T2D, N SNPs = 49), fasting glucose (N SNPs = 36), insulin resistance (N SNPs = 10), body mass index (BMI, N SNPs = 32), total cholesterol (N SNPs = 73), HDL-cholesterol (N SNPs = 71), LDL-cholesterol (N SNPs = 57), triglycerides (N SNPs = 39), systolic blood pressure (SBP, N SNPs = 24), smoking initiation (N SNPs = 1), smoking quantity (N SNPs = 3), university completion (N SNPs = 2), and years of education (N SNPs = 1). We calculated MR estimates of associations between each exposure and AD risk using an inverse-variance weighted approach, with summary statistics of SNP–AD associations from the International Genomics of Alzheimer’s Project, comprising a total of 17,008 individuals with AD and 37,154 cognitively normal elderly controls. We found that genetically predicted higher SBP was associated with lower AD risk (odds ratio [OR] per standard deviation [15.4 mm Hg] of SBP [95% CI]: 0.75 [0.62–0.91]; p = 3.4 × 10−3). Genetically predicted higher SBP was also associated with a higher probability of taking antihypertensive medication (p = 6.7 × 10−8). Genetically predicted smoking quantity was associated with lower AD risk (OR per ten cigarettes per day [95% CI]: 0.67 [0.51–0.89]; p = 6.5 × 10−3), although we were unable to stratify by smoking history; genetically predicted smoking initiation was not associated with AD risk (OR = 0.70 [0.37, 1.33]; p = 0.28). We saw no evidence of causal associations between glycemic traits, T2D, BMI, or educational attainment and risk of AD (all p > 0.1). Potential limitations of this study include the small proportion of intermediate trait variance explained by genetic variants and other implicit limitations of MR analyses.
Conclusions
Inherited lifetime exposure to higher SBP is associated with lower AD risk. These findings suggest that higher blood pressure—or some environmental exposure associated with higher blood pressure, such as use of antihypertensive medications—may reduce AD risk.
Robert A. Scott and colleagues use genetic instruments to identify causal associations between known risk factors and Alzheimer's disease.
Editors' Summary
Background
Worldwide, about 44 million people have dementia, a group of brain degeneration disorders characterized by an irreversible decline in memory, communication, and other “cognitive” functions. Dementia mainly affects older people, and because people are living longer, experts estimate that more than 135 million people will have dementia by 2050. The most common form of dementia, which accounts for 60%–70% of cases, is Alzheimer disease (AD). The earliest sign of AD is often increasing forgetfulness. As the disease progresses, affected individuals gradually lose the ability to look after themselves, they may become anxious or aggressive, and they may have difficulty recognizing friends and relatives. People with late stage disease may lose control of their bladder and of other physical functions. At present, there is no cure for AD, although some of its symptoms can be managed with drugs. Most people with AD are initially cared for at home by relatives and other caregivers, but many affected individuals end their days in a care home or specialist nursing home.
Why Was This Study Done?
Researchers are interested in identifying risk factors for AD, particularly modifiable risk factors, because if such risk factors exist, it might be possible to limit the predicted increase in future AD cases. Epidemiological studies (investigations that examine patterns of disease in populations) have identified several potential risk factors for AD, including hypertension (high blood pressure), obesity, smoking, and dyslipidemia (changes in how the body handles fats). However, epidemiological studies cannot prove that a specific risk factor causes AD. For example, people with hypertension might share another characteristic that causes both hypertension and AD (confounding) or AD might cause hypertension (reverse causation). Information on causality is needed to decide which risk factors to target to help prevent AD. Here, the researchers use “Mendelian randomization” to examine whether differences in several epidemiologically identified risk factors for AD have a causal impact on AD risk. In Mendelian randomization, causal associations are inferred from the effects of genetic variants (which predict levels of modifiable risk factors) on the outcome of interest. Because gene variants are inherited randomly, they are not prone to confounding and are free from reverse causation. So, if hypertension actually causes AD, genetic variants that affect hypertension should be associated with an altered risk of AD.
What Did the Researchers Do and Find?
The researchers identified causal associations between potentially modifiable risk factors and AD risk by analyzing the occurrence of single nucleotide polymorphisms (SNPs, a type of gene variant) known to predict levels of each risk factor, in genetic data from 17,008 individuals with AD and 37,154 cognitively normal elderly controls collected by the International Genomics of Alzheimer’s Project. They report that genetically predicted higher systolic blood pressure (SBP; the pressure exerted on the inside of large blood vessels when the heart is pumping out blood) was associated with lower AD risk (and with a higher probability of taking antihypertensive medication). Predicted smoking quantity was also associated with lower AD risk, but there was no evidence of causal associations between any of the other risk factors investigated and AD risk.
What Do These Findings Mean?
In contrast to some epidemiological studies, these findings suggest that hypertension is associated with lower AD risk. However, because genetically predicted higher SBP was also associated with a higher probability of taking antihypertensive medication, it could be that exposure to such drugs, rather than having hypertension, reduces AD risk. Like all Mendelian randomization studies, the reliability of these findings depends on the validity of several assumptions made by the researchers and on the ability of the SNPs used in the analyses to explain variations in exposure to the various risk factors. Moreover, because all the participants in the International Genomics of Alzheimer’s Project are of European ancestry, these findings may not be valid for other ethnic groups. Given that hypertension is a risk factor for cardiovascular disease, the researchers do not advocate raising blood pressure as a measure to prevent AD (neither do they advocate that people smoke more cigarettes to lower AD risk). Rather, given the strong association between higher SBP gene scores and the probability of exposure to antihypertensive treatment, they suggest that the possibility that antihypertensive drugs might reduce AD risk independently of their effects on blood pressure should be investigated as a priority.
Additional Information
This list of resources contains links that can be accessed when viewing the PDF on a device or via the online version of the article at http://dx.doi.org/10.1371/journal.pmed.1001841.
The UK National Health Service Choices website provides information (including personal stories) about Alzheimer disease
The UK not-for-profit organization Alzheimer’s Society provides information for patients and carers about dementia, including personal experiences of living with Alzheimer disease
The US not-for-profit organization Alzheimer’s Association also provides information for patients and carers about dementia and personal stories about dementia
Alzheimer’s Disease International is the federation of Alzheimer disease associations around the world; it provides links to individual Alzheimer associations, information about dementia, and links to world Alzheimer reports
MedlinePlus provides links to additional resources about Alzheimer disease (in English and Spanish)
Wikipedia has a page on Mendelian randomization (note: Wikipedia is a free online encyclopedia that anyone can edit; available in several languages)
A PLOS Medicine Research Article by Proitsi et al. describes a Mendelian randomization study that looked for a causal association between dyslipidemia and Alzheimer disease
Introduction
Alzheimer disease (AD) prevalence is rising [1], further increasing the social and economic burden of this disease [2]. Epidemiological studies have aimed to identify potentially modifiable risk factors that could be targeted in preventive measures to reduce the incidence of AD. These include type 2 diabetes (T2D) and glycemic traits [3,4], hypertension [5], obesity [6], dyslipidemia [7], smoking [8], physical inactivity [5], depression [9], and low educational attainment [5]. It has been reported that approximately one-third of AD cases worldwide may be attributable to these risk factors [9]. However, this suggestion is predicated on these risk factors having causal effects on AD risk, which is currently uncertain [9]. Given the difficulties in implementing large-scale randomized trials of risk factor modification, alternative approaches are required to investigate the causality of associations and to prioritize the targets for which interventions may be most fruitful [10].
One method for estimating the causal effects of risk factors with known genetic determinants is Mendelian randomization (MR) [11]. The MR approach exploits the fact that genotypes are randomly assorted at meiosis, and are thus independent of conventional confounding factors and the disease process. Therefore, genetic variants associated with intermediate traits can be used to provide an unconfounded estimate of the causal association between the intermediate trait and disease outcome, unaffected by reverse causality. This is akin to a “genetically randomized trial.” For example, if body mass index (BMI) is causally associated with AD, genetic variants causing higher BMI should also be associated with higher risk of AD. However, if an observed BMI—AD association is not causal but is due to confounding or reverse causation, genetic variants causing higher BMI would not result in higher risk of AD. Here, we sought to estimate the causal effects of potentially modifiable risk factors on risk of AD using MR to inform the etiology of AD and the extent to which AD may be preventable by interventions targeting potentially modifiable risk factors.
Methods
Study Design
We performed MR analyses using single nucleotide polymorphisms (SNPs) with known associations with potentially modifiable AD risk factors. We used summary statistics from the International Genomics of Alzheimer’s Project (IGAP) [12], the largest genome-wide meta-analysis of AD reported to date, and individual genotype data from a large subset of IGAP to estimate the unconfounded association between each risk factor and AD risk. S1 Fig illustrates the study design.
SNPs Associated with Alzheimer Disease Risk Factors
We identified SNPs that had genome-wide significant (p < 5 × 10−8) associations with each risk factor using the largest published genome-wide meta-analysis available in individuals of European ancestry. We identified 49 SNPs associated with T2D [13], 36 with fasting glucose [14], and ten with insulin resistance [14,15]. We identified 32 SNPs associated with BMI [16] and 25 associated with systolic blood pressure (SBP) [17]. Given the overlap of SNPs associated with systolic, diastolic, mean arterial, and pulse pressures, we focused on SBP, which had the largest number of associated SNPs [17,18]. We identified 74 SNPs associated with total cholesterol, 71 with high-density lipoprotein (HDL)–cholesterol, 58 with low-density lipoprotein (LDL)–cholesterol, and 40 with triglycerides [19]. We identified one SNP associated with smoking initiation (rs6265 in BDNF; r 2 = 0.74 with the BDNF BMI-associated variant), and three associated with smoking quantity in smokers [20]. We identified two SNPs associated with the probability of completing university and one associated with the number of years of education [21]. We show the SNPs and their associations with their relative traits in S1 Table. Where lead SNPs were not available, we selected a suitable proxy (r 2 > 0.8; except for rs4420638, where the best available proxy was rs6857 [r 2 = 0.46]), as detailed in S1 Table. Within each trait, no SNPs were in linkage disequilibrium (LD) (r 2 < 0.01). No SNPs have been reported to be associated with physical activity levels or depression at p < 5 × 10−8.
Alzheimer Disease Genetic Data
IGAP is a large two-stage study based upon genome-wide association studies (GWASs) of AD in individuals of European ancestry [12]. In stage 1, IGAP used genotyped and imputed data on 7,055,881 SNPs to meta-analyze four previously published GWAS datasets consisting of 17,008 AD cases and 37,154 controls (full details in S1 Text). Further details on the original genetic discovery analyses, including information regarding recruitment and diagnostic assessment as well as analytical approaches to adjust for population structure, are provided in S1 Text or described in detail elsewhere [12]. We extracted individual SNP associations with AD from IGAP’s stage 1 results. Three SNPs (rs850303 for SBP; rs3177928 for total and LDL-cholesterol; rs645040 for triglycerides) were not available (S1 Table), so were excluded from analyses.
Mendelian Randomization Analyses
We used estimated SNP—risk factor and SNP—AD associations to calculate estimates of each risk factor—AD association using an inverse-variance weighted combination of estimates from each SNP [22]. For continuous exposures (BMI, fasting glucose, insulin resistance, lipids, and SBP), we scaled MR estimates per standard deviation (SD) difference of the risk factor. Effect sizes on log-fasting insulin were used as weights for the insulin-resistance-associated variants. SDs were estimated from up to 10,445 (N min = 9,963) middle-aged adults from the UK population-based Fenland study [23]. Causal estimates are thus presented per genetically predicted SD, and a log-linear association with odds of AD is implicit across the range of intermediate risk factor values. We scaled smoking quantity per ten cigarettes per day and scaled educational attainment per year of education. For binary exposures (T2D, smoking initiation, completing university), MR estimates are odds ratios (ORs) per genetically predicted unit difference in log-odds of having the relevant exposure. Overall, we included 302 non-overlapping SNPs. To minimize the possibility of pleiotropic associations influencing results, we performed sensitivity analyses excluding SNPs with a more significant association with AD than expected by chance (p < 0.05/302 = 0.00017), which excluded only four variants in total (S1 Table). Furthermore, we investigated the association of each variant with the risk factor relative to the magnitude of association with AD risk to further identify variants that appeared to be outliers and were candidates to be pleiotropic. As a further sensitivity analysis, for risk factors that showed evidence of a causal association with AD (p < 3.8 × 10−3), we also performed a “leave one out” analysis to further investigate the possibility that the causal association was driven by a single SNP.
We also performed MR analyses of risk factors that showed evidence of a causal association with AD (p < 3.8 × 10−3) using individual-level SNP data from studies in the Alzheimer’s Disease Genetics Consortium (ADGC) (cases = 10,079; controls = 9,613) [24] and the Genetic and Environmental Risk in AD (GERAD1) Consortium (cases = 3,146; controls = 1,224) [25], which account for 51% of the IGAP effective sample size (see S1 Text for a description of the ADGC and GERAD1 samples). We performed logistic regression analyses of the SNP-predicted AD association adjusting for study site, population substructure, age, and sex, again scaled per 1-SD difference in risk factor.
We created unweighted genetic scores based on the number of risk alleles for each SNP—risk factor association and investigated the association of these scores with a range of traits in up to 16,554 individuals from the EPIC-InterAct study [26] to check the assumption that the SNPs used in the MR analyses are not associated with potential confounders of exposure—AD associations. We standardized outcomes and included scores in linear regression models adjusted for age, sex, recruitment center, and subcohort status. We natural-log-transformed triglyceride levels before standardization. We investigated the association of the SBP-associated variants with both SBP and diastolic blood pressure (DBP). We did not adjust observed blood pressure values for antihypertensive usage. We used logistic regression to determine associations with the probability of being physically active, being a smoker, or taking antihypertensive medications, and included covariates as above. The distribution of the SBP risk score in the EPIC-InterAct study [26] is shown in S2 Fig.
Results
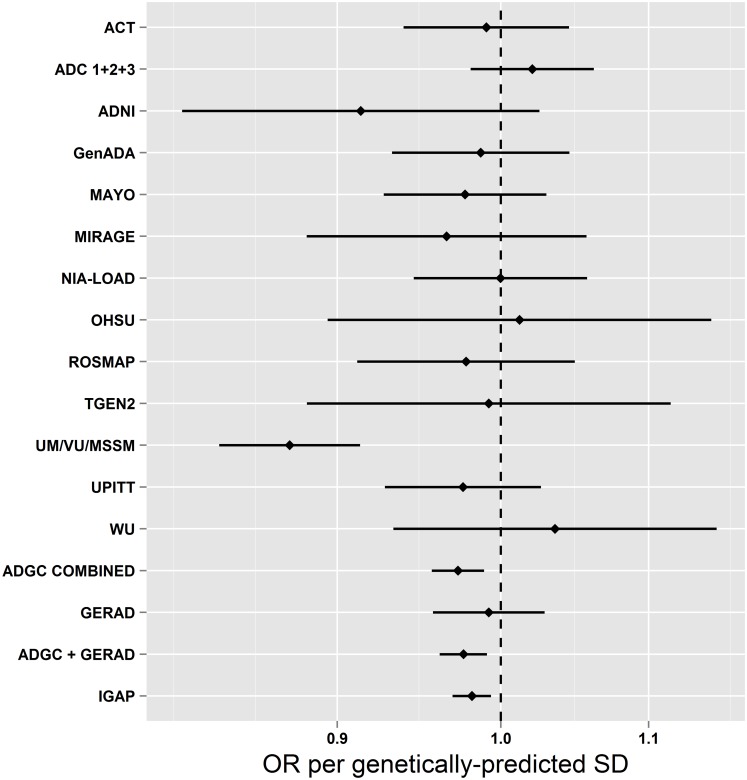
Table 1 shows the estimated associations of each genetically predicted risk factor with AD from MR analysis using a large-scale international investigation of the genetic basis of AD risk in 17,008 individuals with AD and 37,154 controls. We observed evidence for a causal association between genetically predicted SBP and AD risk. A genetically predicted 1-SD (15.4 mm Hg) higher SBP was associated with lower risk of AD (OR [95% CI]: 0.75 [0.62–0.91]; p = 3.4 × 10−3). We examined each of the SBP SNPs to investigate if particular SNPs were driving the association with AD, but observed no obvious outliers (S3 Fig). Furthermore, when we performed all 24 permutations of the “leave one out” analysis, all SNP sets showed consistent evidence of causality (OR per SD of SBP [95% CI] ranged from 0.72 [0.59–0.87] to 0.78 [0.64–0.95]). Individual SNP associations with AD are shown in S1 Table. We also performed analyses on a subset of the overall sample using individual-level SNP data from ADGC and GERAD1, which showed results similar to those observed using the inverse-variance weighted approach (OR [95% CI]: 0.69 [0.55–0.85]; p = 2.0 × 10−3; Fig 1). We saw no evidence of heterogeneity between individual studies (p = 0.33).
Table 1. Estimated associations of each genetically predicted risk factor with Alzheimer disease.
| Trait | Scaling of OR | Number of SNPs | Overall Results | Sensitivity Analyses* | ||
|---|---|---|---|---|---|---|
| OR (95% CI) | p-Value | OR (95% CI) | p-Value | |||
| BMI | 1 SD (4.81 kg/m2) | 32 | 0.99 (0.80−1.19) | 0.779 | 1.00 (0.82−1.22) | 0.97 |
| T2D | 1 unit higher log-odds | 49 | 1.02 (0.97−1.07) | 0.535 | ||
| Fasting glucose | 1 SD (0.65 mmol/l) | 36 | 1.12 (0.97−1.30) | 0.112 | 1.19 (1.03−1.37) | 0.02 |
| Insulin resistance | 1 SD log-FI (0.60 log-pmol/l) | 10 | 1.32 (0.88−1.98) | 0.177 | ||
| SBP | 1 SD (15.4 mm Hg) | 24 | 0.75 (0.62−0.91) | 3.4 × 10−3 | ||
| Total cholesterol | 1 SD (1.03 mmol/l) | 73 | 1.94 (1.79−2.10) | 3.1 × 10−56 | 1.04 (0.95−1.13) | 0.84 |
| HDL-cholesterol | 1 SD (0.41 mmol/l) | 71 | 0.75 (0.69−0.82) | 1.0 × 10−11 | 1.01 (0.93−1.09) | 0.87 |
| LDL-cholesterol | 1 SD (0.91 mmol/l) | 57 | 2.31 (2.12−2.50) | 3.0 × 10−87 | 1.07 (0.98−1.17) | 0.14 |
| Triglycerides | 1 SD (0.83 mmol/l) | 39 | 0.96 (0.87−1.07) | 0.482 | ||
| Smoking initiation | 1 unit higher log-odds | 1 | 0.70 (0.37−1.33) | 0.278 | ||
| Smoking quantity | 10 cigarettes/day | 3 | 0.67 (0.51−0.89) | 6.5 × 10−3 | ||
| Completing university | 1 unit higher log-odds | 2 | 0.95 (0.67−1.34) | 0.752 | ||
| Length of education | 1 year of education | 1 | 0.71 (0.48−1.06) | 0.097 | ||
*Sensitivity analyses exclude SNPs where p < 0.00017 (0.05/302 unique SNPs) for AD.
log-FI, log-fasting insulin.
Fig 1. Mendelian randomization estimates of the association of systolic blood pressure with AD in individual ADGC studies and overall in ADGC, GERAD1, and IGAP.
This figure shows MR estimates for the association of SBP-associated variants with AD in each of the participant studies in ADGC [24] and in GERAD1 [25] using individual SNP-level data compared to that observed in IGAP [12] using summary-level data. See S1 Text (supplemental results) for individual study name abbreviations.
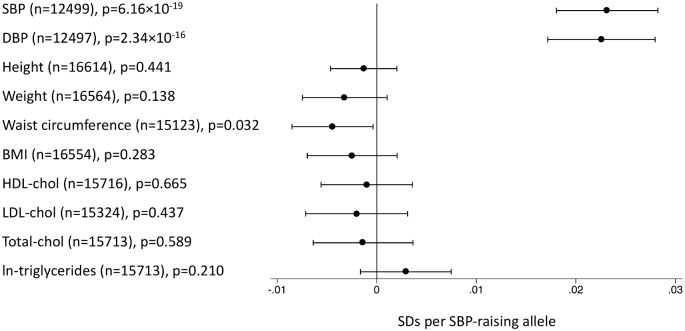
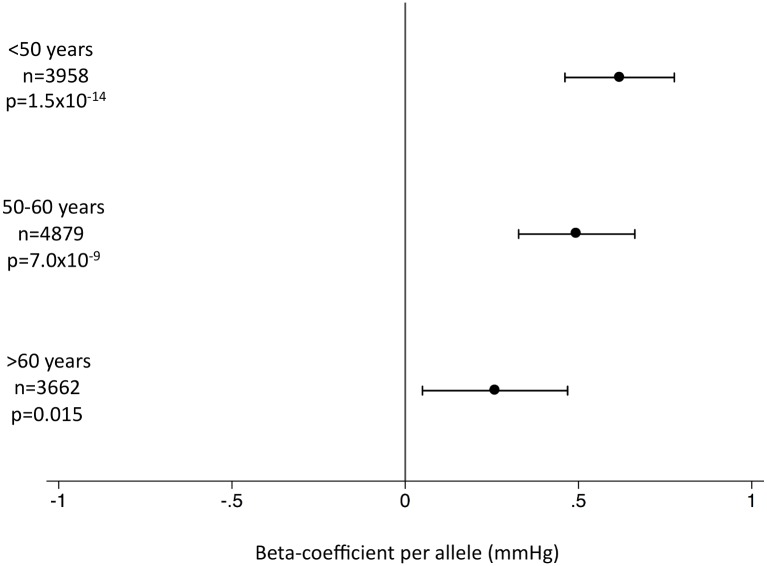
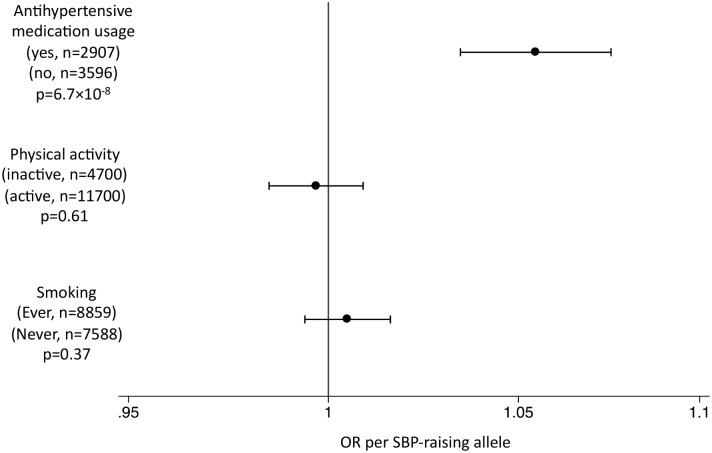
In the EPIC-InterAct study, the unweighted SBP genetic score was strongly associated with SBP and DBP overall (Fig 2) and in all age groups (Fig 3) (p < 0.015 for SBP; p < 0.002 for DBP). We did not observe associations of the SBP score with other potentially confounding variables in the EPIC-InterAct study (Fig 2). The unweighted SBP genetic score was associated with a higher probability of taking antihypertensive medication (OR [95% CI]: 1.05 [1.03–1.08]; p = 6.7 × 10−8) but not with the probability of being physically active or being a smoker (Fig 4). Forty-nine percent of the individuals in the highest quartile of the unweighted SBP genetic score reported taking antihypertensive medication compared to 39% in the lowest quartile.
Fig 2. Associations of the systolic blood pressure genetic score with quantitative traits in the EPIC-InterAct study.
This figure shows the investigation of pleiotropic associations of genetic score for SBP with quantitative traits in the EPIC-InterAct study [26]. Effect sizes are expressed in SDs per SBP-raising allele. Analyses were adjusted for age, sex, center of recruitment, and subcohort status.
Fig 3. Association of the systolic blood pressure genetic score with systolic blood pressure by age stratum in the EPIC-InterAct subcohort.
This figure shows the association between the genetic score for SBP and SBP in the EPIC-InterAct study by age stratum [26]. Analyses were adjusted for sex, center of recruitment, and subcohort status.
Fig 4. Associations of the systolic blood pressure genetic score with binary outcomes in the EPIC-InterAct study.
This figure shows the investigation of pleiotropic associations of the genetic score for SBP with binary outcomes in the EPIC-InterAct study [26]. The OR per SBP-raising allele is shown.
We found strong associations between genetically predicted total, LDL-, and HDL-cholesterol and AD (Table 1). Each of these SNP sets included rs6857 near APOE, which is strongly associated with AD risk (OR: 3.2; p = 2.5 × 10−575) [12] (S4 and S5 Figs) and which was a very clear outlier when we compared effect sizes on lipids against effect sizes on AD (S6–S11 Figs). Two of these SNP sets also included rs1883025 from ABCA1, which was associated with AD at a significance level beyond that expected by chance (OR: 1.07; p = 1.0 × 10−4) [12] and which is in a gene previously implicated in association with AD [27]. After sensitivity analyses excluding these potentially pleiotropic SNPs, we saw no evidence for causal associations between lipid fractions and AD risk (Table 1).
We found no evidence to support causal associations between BMI and AD (OR per SD of BMI [95% CI]: 0.99 [0.80–1.19]; p = 0.78), fasting glucose (OR [95% CI]:1.12 [0.97–1.30]; p = 0.11), insulin resistance (OR [95% CI]: 1.32 [0.89–1.97]; p = 0.17), or T2D (OR [95% CI]: 1.01 [0.96–1.07]; p = 0.57) (Table 1). S4 Fig shows the associations with AD of the SNPs included in all genetic analyses compared to those expected by chance. Other than rs6857 near APOE, the most significant associations with AD were observed for rs11039149 (p = 3.7 × 10−6) in the fasting glucose SNPs and rs3817334 (p = 9.3 × 10−5) in the BMI SNPs (see S1 Table and S5 Fig). These SNPs are both in LD (r 2 = 0.58 and r 2 = 0.33, respectively) with a genome-wide significant association signal for AD in CELF1 [12]. After excluding these variants from their respective SNP sets, BMI results were unchanged (Table 1). However, for fasting glucose, following the removal of rs11039149 near MADD, there was a suggestion of an association between higher glucose and higher AD risk (OR [95% CI]: 1.19 [1.03–1.37]; p = 0.02).
We found no evidence to support causal associations between smoking initiation and AD (OR [95% CI]: 0.70 [0.37–1.33]; p = 0.28). We did find an association between genetically predicted higher smoking quantity and lower AD (OR per ten cigarettes/day [95% CI]: 0.67 [0.51–0.89]; p = 6.5 × 10−3). We did not have smoking behavior data for IGAP to obtain estimates for the association with AD among smokers and non-smokers. The SNP with the strongest association with smoking quantity [20] was nominally associated with AD risk (rs1051730: OR of AD per smoking-quantity-raising allele [95% CI]: 0.96 [0.93–0.99]; p = 0.01), while the others were not (S12 Fig). We saw no association between AD risk and either university completion (OR [95% CI]: 0.95 [0.67–1.34]; p = 0.75) or years of education (OR [95% CI]: 0.71 [0.48–1.06]; p = 0.10) (Table 1).
Discussion
The potential of risk factor modifications to impact upon AD incidence depends entirely on causal links between the risk factors and AD. Using genetic variants associated with risk factors for AD in a very large consortium of well-characterized research participants, we found evidence for an association between genetically inherited higher levels of blood pressure and lower AD risk.
Hypertension has been implicated as a risk factor for AD [5]. However, uncertainties remain over the nature of the association, perhaps complicated by misclassification of AD with other forms of dementia, or the age of study participants [28]. While previous studies have suggested that high blood pressure in midlife is associated with higher AD risk [29,30], other studies have indicated that high blood pressure in late life may be protective against AD [31,32]. We found that genetically inherited higher SBP levels are associated with lower risk of AD (Table 1). Previous studies have suggested that hypotension may indeed be a risk factor for AD, particularly in the elderly [33], potentially via resultant cerebral hypoperfusion [34]. The unweighted SBP gene score was associated with higher SBP levels across the adult lifespan (Fig 3). It should be noted that the SNPs associated with SBP overlap extensively with those associated with DBP [17] as well as with pulse pressure [18], so we were unable to distinguish between individual components of blood pressure. A recent meta-analysis of prospective studies suggested that a 10-mm Hg higher SBP was associated with a protective relative risk of 0.95 (95% CI: 0.91–1.00) for AD [28]. Scaling our results to a genetically predicted 10-mm Hg difference in SBP would result in an OR of 0.83 (95% CI: 0.73–0.94) for AD. Clearly, given that blood pressure is a major risk factor for cardiovascular disease [35], one would not advocate raising blood pressure as a preventive strategy, yet these findings offer intriguing etiological insight.
We also found that genetically predicted higher SBP was associated with a higher probability of being on antihypertensive medication (Fig 4). There is considerable interest in the role of antihypertensives in dementia, and while findings are equivocal [36], recent studies have suggested a possible protective effect of antihypertensive therapy on AD risk [37], potentially with heterogeneity of effect by therapeutic class [38], suggesting that any effect on AD risk may not be entirely attributable to the lowering of blood pressure, but potentially to other mechanisms. The unweighted SBP gene score was strongly associated with observed SBP in the EPIC-InterAct study, ignoring any SBP-lowering effects of antihypertensive medications. Thus, if antihypertensive medications are indeed protective and confound the association between genetically predicted SBP and AD, their effect on AD risk is likely to be independent of their effect on SBP, as the SBP-associated variants have a strong association with SBP regardless of the higher prevalence of treatment with antihypertensive medication. While the null association between genetically predicted lipid levels and AD risk reflects the equivocal findings from trials of statins and cognitive decline [39], our results suggest the imperative need for further investigation of the possibility that antihypertensive medications may reduce AD risk independently of their effects on blood pressure. Future MR analyses stratified by antihypertensive treatments would be desirable to more precisely estimate the magnitude of the causal effect of higher BP on AD risk, but will be difficult to carry out using existing data due to the time-varying nature of antihypertensive treatments across the life course, and the non-availability of data on lifetime medication usage in most studies.
We also observed an association between AD and smoking quantity (Table 1). Early reports implicated smoking as protective for AD [40], potentially via a neuroprotective effect of nicotine [41]. However, this association may be due to differential survival bias [42], and a recent meta-analysis of prospective studies implicates smoking as a risk factor for AD, showing current smokers as being at higher risk of AD than never smokers [8]. One smoking-quantity-related SNP was associated with AD (p = 0.01). This SNP is in the gene CHRNA3 from the nicotinic receptor gene cluster CHRNA5-CHRNA3-CHRNB4. Given the putative actions of nicotine, variants in this locus may confer neuroprotective effects by influencing nicotinic receptor function [41,43]. Thus, altered nicotinic receptor function may underlie the MR association between smoking quantity and AD risk. The ideal study would perform MR analyses stratified by smoking status [44], particularly if sensitivity analyses could exclude variants in nicotinic receptor genes. Such analyses would address the causality of smoking as a risk factor, and offer valuable insight into nicotine’s role in the etiology of AD [41,43]. Since smoking is a major cause of global disease burden [45], increasing knowledge of the role of nicotine in the etiology of AD may prove to be the more actionable insight.
Our findings for total, LDL-, and HDL-cholesterol are not consistent with a causal effect of major lipid fractions on AD risk, as previously suggested in a smaller study [46]. Rather, the well-established association of APOE haplotypes with AD risk [47] implicates APOE itself as a key causal factor in the etiology of AD. Indeed a recent GWAS of plasma APOE levels identified only genetic variants in APOE, and not those in other lipid loci, as being associated with APOE levels at genome-wide significance [48]. When we compared the effect sizes for the effects of SNPs on major lipids relative to the magnitude of their association with AD, the APOE variant was a very clear outlier (S6–S11 Figs).
We did not find evidence consistent with a causal role for the other potentially modifiable risk factors we evaluated (Table 1). In our sensitivity analysis that excluded the potentially pleiotropic variant near MADD, genetically predicted higher fasting glucose was nominally associated with higher AD risk. While these results are consistent with the notion that higher blood glucose may be causally related to AD risk [4], the borderline significance warrants a cautious interpretation.
A limitation of the MR approach is the limited strength of the SNPs to explain variation in the intermediate traits, restricting statistical power. This is particularly true when findings are null, where narrow confidence intervals are important to aid robust inference. For example, while we saw no evidence to support causal roles for BMI, fasting glucose, or insulin resistance in AD (all p > 0.1), confidence intervals allow for an almost 20% higher AD risk per 1-SD difference in BMI, a 30% higher AD risk per 1-SD difference in fasting glucose, and an almost 100% higher AD risk per 1-SD difference in log-fasting insulin (Table 1). Thus, improving the intermediate trait variance explained by the instrumental variables by further genetic discovery efforts will improve the precision of MR analyses. Likewise, ever larger AD GWASs will further narrow confidence intervals around MR estimates. The association of genetically predicted blood pressure with AD risk remained after Bonferroni correction for the 13 individual SNP sets we tested (0.05/13 = 3.8 × 10−3), although the association of the smoking-associated variants did not. However, we consider this a conservative correction, given the correlation between the intermediate risk factors. We cannot exclude the possibility that the protective associations of blood pressure with AD arise as a result of differential survival bias, but the consistency of the observations across both prospective and cross-sectional studies of AD makes this less likely (Fig 1), as does the absence of similar MR associations for other major vascular risk factors (Table 1).
The main data source for this study is the summary statistics from IGAP, the largest genome-wide meta-analysis of AD reported to date [12]. Since all participants in IGAP are of European ancestry, the results of this study are not necessarily valid for other ethnic groups.
In conclusion, we found associations between genetically predicted higher SBP and lower AD risk. This finding is contrary to the notion that societal interventions to lower blood pressure will reduce the incidence of AD. However, since there is a strong association between higher SBP gene scores and exposure to antihypertensive treatments, there is a need to evaluate the possible protective role of some of these substances against AD, independent of their effects on blood pressure.
Supporting Information
(DOCX)
(TIF)
n = 16,691.
(TIF)
(TIF)
SNPs from all scores (N unique = 302) were LD-pruned and duplicates were removed to leave the 269 variants shown here. The SNP near APOE (rs6857) had a p-value of 2.5 × 10−575, which was truncated to 10−30 for display on the figure.
(TIF)
SNPs from all scores (N unique = 302) were LD-pruned and duplicates were removed to leave 269 variants. For the present plot, the SNP near APOE (rs6857, p = 2.5 × 10−575) was excluded (see S4 Fig).
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
*Note that the effect sizes for the effects of SNPs on smoking quantity were estimated in current smokers only, while the associations with AD are not stratified by smoking status.
(TIF)
SNPs included in each risk score, their associations estimated by the relevant consortium, and their association with AD (IGAP [12]). Proxies are marked by an asterix in the leftmost column. The effect allele frequency was based on data from the EPIC-InterAct study [26]. Double asterisks indicate that frequency information was not available in the EPIC-InterAct study, and were obtained from European 1000 Genomes samples.
(XLSX)
(DOCX)
Acknowledgments
See S1 Text.
Abbreviations
- AD
Alzheimer disease
- ADGC
Alzheimer’s Disease Genetics Consortium
- BMI
body mass index
- DBP
diastolic blood pressure
- GERAD1
Genetic and Environmental Risk in AD
- GWAS
genome-wide association study
- HDL
high-density lipoprotein
- IGAP
International Genomics of Alzheimer’s Project
- LD
linkage disequilibrium
- LDL
low-density lipoprotein
- MR
Mendelian randomization
- OR
odds ratio
- SD
standard deviation
- SNP
single nucleotide polymorphism
- SBP
systolic blood pressure
- T2D
type 2 diabetes
Data Availability
All association data with Alzheimer's disease for each individual SNP is available from http://www.pasteur-lille.fr/en/recherche/u744/igap/igap_download.php. Individual-level data can be sought by request to the GERAD consortium (http://gtr.rcuk.ac.uk/project/B6C58A7C-3C3E-41CB-AF10-16DB59962C9E). To promote collaboration and wide use of data in order to further Alzheimer’s Disease research, qualified investigators can submit proposals to conduct analyses using results from the Genetic and Environmental Risk in Alzheimer’s Disease (GERAD) analysis or, in collaboration with GERAD, to conduct analyses using data from GERAD members. The PI and other key personnel must be qualified investigators with appropriate experience and have the support of at least one GERAD collaborator. Proposals can be submitted at any time and will be provided to all GERAD members, who will be asked to review and feedback to the GERAD coordinator (Julie Williams) within 2 weeks. Where objections or additional information is sought the proposal may be discussed on the next GERAD conference call. Further revision or clarification may be requested at this time. After all questions are addressed, the GERAD consortium will provide any objections to the coordinator (Julie Williams). All objections will be taken into account and acted on appropriately. Individual members can opt-out of projects. Proposals should not overlap with analyses that are being performed within GERAD, and should be new and novel research ideas. In order to access GERAD data, the receiving Institution is required to enter into a data sharing agreement with Cardiff University which binds them to appropriate use and acknowledgement of the data they receive. Only parties involved in this agreement can access GERAD data for the purpose specified in the proposal.
Funding Statement
This study was supported by the Innovative Medicines Initiative Joint Undertaking under EMIF grant agreement n° 115372 (contributions from the European Union's Seventh Framework Programme (FP7/2007-2013) and EFPIA companies). The work was further supported by the National Institute for Health Research (NIHR) Mental Health Biomedical Research Centre and Dementia Unit at South London and Maudsley NHS Foundation Trust and [Institute of Psychiatry] King’s College London. SDØ is supported by a grant from the Lundbeck Foundation. PP is an Alzheimer’s Society Post-Doctoral Fellow. NJW is an NIHR Senior Investigator. Funding for the EPIC-InterAct project was provided by the EU FP6 programme (grant number LSHM_CT_2006_037197). ADGC funding: The National Institutes of Health, National Institute on Aging (NIH-NIA) supported this work through the following grants: ADGC, U01 AG032984, RC2 AG036528; NACC, U01 AG016976; NCRAD, U24 AG021886; NIA LOAD, U24 AG026395, U24 AG026390; Banner Sun Health Research Institute P30 AG019610; Boston University, P30 AG013846, U01 AG10483, R01 CA129769, R01 MH080295, R01 AG017173, R01 AG025259, R01AG33193; Columbia University, P50 AG008702, R37 AG015473; Duke University, P30 AG028377, AG05128; Emory University, AG025688; Group Health Research Institute, U01 AG06781, U01 HG004610, U01 HG006375; Indiana University, P30 AG10133; Johns Hopkins University, P50 AG005146, R01 AG020688; Massachusetts General Hospital, P50 AG005134; Mayo Clinic, P50 AG016574; Mount Sinai School of Medicine, P50 AG005138, P01 AG002219; New York University, P30 AG08051, MO1RR00096, UL1 RR029893, 5R01AG012101, 5R01AG022374, 5R01AG013616, 1RC2AG036502, 1R01AG035137; Northwestern University, P30 AG013854; Oregon Health & Science University, P30 AG008017, R01 AG026916; Rush University, P30 AG010161, R01 AG019085, R01 AG15819, R01 AG17917, R01 AG30146; TGen, R01 NS059873; University of Alabama at Birmingham, P50 AG016582, UL1RR02777; University of Arizona, R01 AG031581; University of California, Davis, P30 AG010129; University of California, Irvine, P50 AG016573, P50 AG016575, P50 AG016576, P50 AG016577; University of California, Los Angeles, P50 AG016570; University of California, San Diego, P50 AG005131; University of California, San Francisco, P50 AG023501, P01 AG019724; University of Kentucky, P30 AG028383, AG05144; University of Michigan, P50 AG008671; University of Pennsylvania, P30 AG010124; University of Pittsburgh, P50 AG005133, AG030653, AG041718; University of Southern California, P50 AG005142; University of Texas Southwestern, P30 AG012300; University of Miami, R01 AG027944, AG010491, AG027944, AG021547, AG019757; University of Washington, P50 AG005136; Vanderbilt University, R01 AG019085; and Washington University, P50 AG005681, P01 AG03991. The Kathleen Price Bryan Brain Bank at Duke University Medical Center is funded by NINDS grant # NS39764, NIMH MH60451 and by Glaxo Smith Kline. Genotyping of the TGEN2 cohort was supported by Kronos Science. The TGen series was also funded by NIA grant AG041232 to AJM and MJH, The Banner Alzheimer’s Foundation, The Johnnie B. Byrd Sr. Alzheimer’s Institute, the Medical Research Council, and the state of Arizona and also includes samples from the following sites: Newcastle Brain Tissue Resource (funding via the Medical Research Council, local NHS trusts and Newcastle University), MRC London Brain Bank for Neurodegenerative Diseases (funding via the Medical Research Council),South West Dementia Brain Bank (funding via numerous sources including the Higher Education Funding Council for England (HEFCE), Alzheimer’s Research Trust (ART), BRACE as well as North Bristol NHS Trust Research and Innovation Department and DeNDRoN), The Netherlands Brain Bank (funding via numerous sources including Stichting MS Research, Brain Net Europe, Hersenstichting Nederland Breinbrekend Werk, International Parkinson Fonds, Internationale Stiching Alzheimer Onderzoek), Institut de Neuropatologia, Servei Anatomia Patologica, Universitat de Barcelona. Funding for ADNI is through the Northern California Institute for Research and Education by grants from Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson and Johnson, Eli Lilly and Co., Medpace, Inc., Merck and Co., Inc., Novartis AG, Pfizer Inc, F. Hoffman-La Roche, Schering-Plough, Synarc, Inc., Alzheimer's Association, Alzheimer's Drug Discovery Foundation, the Dana Foundation, and by the National Institute of Biomedical Imaging and Bioengineering and NIA grants U01 AG024904, RC2 AG036535, K01 AG030514. We thank Drs. D. Stephen Snyder and Marilyn Miller from NIA who are ex-officio ADGC members. Support was also from the Alzheimer’s Association (LAF, IIRG-08-89720; MP-V, IIRG-05-14147) and the US Department of Veterans Affairs Administration, Office of Research and Development, Biomedical Laboratory Research Program. P.S.G.-H. is supported by Wellcome Trust, Howard Hughes Medical Institute, and the Canadian Institute of Health Research. ACT is supported by a grant (U01 AG 06781, to Dr. Larson) from the National Institutes of Health. GERAD1 funding: Cardiff University was supported by the Wellcome Trust, Medical Research Council (MRC), Alzheimer’s Research UK (ARUK) and the Welsh Assembly Government. Cambridge University and Kings College London acknowledge support from the MRC. ARUK supported sample collections at the South West Dementia Bank and the Universities of Nottingham, Manchester and Belfast. The Belfast group acknowledges support from the Alzheimer's Society, Ulster Garden Villages, N.Ireland R&D Office and the Royal College of Physicians/Dunhill Medical Trust. The MRC and Mercer’s Institute for Research on Ageing supported the Trinity College group. The South West Dementia Brain Bank acknowledges support from Bristol Research into Alzheimer's and Care of the Elderly. The Charles Wolfson Charitable Trust supported the OPTIMA group. Washington University was funded by NIH grants, Barnes Jewish Foundation and the Charles and Joanne Knight Alzheimer's Research Initiative. Patient recruitment for the MRC Prion Unit/UCL Department of Neurodegenerative Disease collection was supported by the UCLH/UCL Biomedical Centre and NIHR Queen Square Dementia Biomedical Research Unit. LASER-AD was funded by Lundbeck SA. The Bonn group was supported by the German Federal Ministry of Education and Research (BMBF), Competence Network Dementia and Competence Network Degenerative Dementia, and by the Alfried Krupp von Bohlen und Halbach-Stiftung. The GERAD Consortium also used samples ascertained by the NIMH AD Genetics Initiative. The KORA F4 studies were financed by Helmholtz Zentrum München; German Research Center for Environmental Health; BMBF; German National Genome Research Network and the Munich Center of Health Sciences. The Heinz Nixdorf Recall cohort was funded by the Heinz Nixdorf Foundation (Dr. jur. G.Schmidt, Chairman) and BMBF. Coriell Cell Repositories is supported by NINDS and the Intramural Research Program of the National Institute on Aging. We acknowledge use of genotype data from the 1958 Birth Cohort collection, funded by the MRC and the Wellcome Trust which was genotyped by the Wellcome Trust Case Control Consortium and the Type-1 Diabetes Genetics Consortium, sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Allergy and Infectious Diseases, National Human Genome Research Institute, National Institute of Child Health and Human Development and Juvenile Diabetes Research Foundation International. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Prince M, Bryce R, Albanese E, Wimo A, Ribeiro W, Ferri CP. The global prevalence of dementia: a systematic review and metaanalysis. Alzheimers Dement. 2013;9:63–75. e2 10.1016/j.jalz.2012.11.007 [DOI] [PubMed] [Google Scholar]
- 2. Reitz C, Mayeux R. Alzheimer disease: epidemiology, diagnostic criteria, risk factors and biomarkers. Biochem Pharmacol. 2014;88:640–651. 10.1016/j.bcp.2013.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vagelatos NT, Eslick GD. Type 2 diabetes as a risk factor for Alzheimer’s disease: the confounders, interactions, and neuropathology associated with this relationship. Epidemiol Rev. 2013;35:152–160. 10.1093/epirev/mxs012 [DOI] [PubMed] [Google Scholar]
- 4. Crane PK, Walker R, Hubbard RA, Li G, Nathan DM, Zheng H, et al. Glucose levels and risk of dementia. N Engl J Med. 2013;369:540–548. 10.1056/NEJMoa1215740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barnes DE, Yaffe K. The projected effect of risk factor reduction on Alzheimer’s disease prevalence. Lancet Neurol. 2011;10:819–828. 10.1016/S1474-4422(11)70072-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Anstey KJ, Cherbuin N, Herath PM. Development of a new method for assessing global risk of Alzheimer’s disease for use in population health approaches to prevention. Prev Sci. 2013;14:411–421. 10.1007/s11121-012-0313-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Whitmer RA, Sidney S, Selby J, Johnston SC, Yaffe K. Midlife cardiovascular risk factors and risk of dementia in late life. Neurology. 2005;64:277–281. [DOI] [PubMed] [Google Scholar]
- 8. Peters R, Poulter R, Warner J, Beckett N, Burch L, Bulpitt C. Smoking, dementia and cognitive decline in the elderly, a systematic review. BMC Geriatr. 2008;8:36 10.1186/1471-2318-8-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Norton S, Matthews FE, Barnes DE, Yaffe K, Brayne C. Potential for primary prevention of Alzheimer’s disease: an analysis of population-based data. Lancet Neurol. 2014;13:788–794. 10.1016/S1474-4422(14)70136-X [DOI] [PubMed] [Google Scholar]
- 10. Burgess S, Butterworth A, Malarstig A, Thompson SG. Use of Mendelian randomisation to assess potential benefit of clinical intervention. BMJ. 2012;345:e7325 10.1136/bmj.e7325 [DOI] [PubMed] [Google Scholar]
- 11. Davey Smith G, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. [DOI] [PubMed] [Google Scholar]
- 12. Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. 10.1038/ng.2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. 10.1038/ng.2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Scott RA, Lagou V, Welch RP, Wheeler E, Montasser ME, Luan J, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet. 2012;44:991–1005. 10.1038/ng.2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Scott RA, Fall T, Pasko D, Barker A, Sharp SJ, Arriola L, et al. Common genetic variants highlight the role of insulin resistance and body fat distribution in type 2 diabetes, independently of obesity. Diabetes. 2014;63:4378–4387. 10.2337/db14-0319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. 10.1038/ng.686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. International Consortium for Blood Pressure Genome-Wide Association Studies, Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. 10.1038/nature10405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wain LV, Verwoert GC, O’Reilly PF, Shi G, Johnson T, Johnson AD, et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat Genet. 2011;43:1005–1011. 10.1038/ng.922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Global Lipids Genetics Consortium, Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. 10.1038/ng.2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–447. 10.1038/ng.571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rietveld CA, Medland SE, Derringer J, Yang J, Esko T, Martin NW, et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science. 2013;340:1467–1471. 10.1126/science.1235488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658–665. 10.1002/gepi.21758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rolfe Ede L, Loos RJ, Druet C, Stolk RP, Ekelund U, Griffin SJ, et al. Association between birth weight and visceral fat in adults. Am J Clin Nutr. 2010;92:347–352. 10.3945/ajcn.2010.29247 [DOI] [PubMed] [Google Scholar]
- 24. Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43:436–441. 10.1038/ng.801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. 10.1038/ng.440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. InterAct Consortium, Langenberg C, Sharp S, Forouhi NG, Franks PW, Schulze MB, et al. Design and cohort description of the InterAct project: an examination of the interaction of genetic and lifestyle factors on the incidence of type 2 diabetes in the EPIC study. Diabetologia. 2011;54:2272–2282. 10.1007/s00125-011-2182-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Katzov H, Chalmers K, Palmgren J, Andreasen N, Johansson B, Cairns NJ, et al. Genetic variants of ABCA1 modify Alzheimer disease risk and quantitative traits related to beta-amyloid metabolism. Hum Mutat. 2004;23:358–367. [DOI] [PubMed] [Google Scholar]
- 28. Power MC, Weuve J, Gagne JJ, McQueen MB, Viswanathan A, Blacker D. The association between blood pressure and incident Alzheimer disease: a systematic review and meta-analysis. Epidemiology. 2011;22:646–659. 10.1097/EDE.0b013e31822708b5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Launer LJ, Ross GW, Petrovitch H, Masaki K, Foley D, White LR, et al. Midlife blood pressure and dementia:the Honolulu-Asia aging study. Neurobiol Aging. 2000;21:49–55. [DOI] [PubMed] [Google Scholar]
- 30. Kivipelto M, Helkala EL, Laakso MP, Hanninen T, Hallikainen M, Alhainen K, et al. Midlife vascular risk factors and Alzheimer’s disease in later life: longitudinal, population based study. BMJ. 2001;322:1447–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Qiu C, Winblad B, Fratiglioni L. Low diastolic pressure and risk of dementia in very old people: a longitudinal study. Dement Geriatr Cogn Disord. 2009;28:213–219. 10.1159/000236913 [DOI] [PubMed] [Google Scholar]
- 32. Albanese E, Lombardo FL, Prince MJ, Stewart R. Dementia and lower blood pressure in Latin America, India, and China: a 10/66 cross-cohort study. Neurology. 2013;81:228–235. 10.1212/WNL.0b013e31829bfe66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guo Z, Viitanen M, Fratiglioni L, Winblad B. Low blood pressure and dementia in elderly people: the Kungsholmen project. BMJ. 1996;312:805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de la Torre JC. Cardiovascular risk factors promote brain hypoperfusion leading to cognitive decline and dementia. Cardiovasc Psychiatry Neurol. 2012;2012:367516 10.1155/2012/367516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lewington S, Clarke R, Qizilbash N, Peto R, Collins R, Prospective Studies Collaboration. Age-specific relevance of usual blood pressure to vascular mortality: a meta-analysis of individual data for one million adults in 61 prospective studies. Lancet. 2002;360:1903–1913. [DOI] [PubMed] [Google Scholar]
- 36. Forette F, Seux ML, Staessen JA, Thijs L, Birkenhager WH, Babarskiene MR, et al. Prevention of dementia in randomised double-blind placebo-controlled systolic hypertension in Europe (Syst-Eur) trial. Lancet. 1998;352:1347–1351. [DOI] [PubMed] [Google Scholar]
- 37. Yasar S, Xia J, Yao W, Furberg CD, Xue QL, Mercado CI, et al. Antihypertensive drugs decrease risk of Alzheimer disease: Ginkgo Evaluation of Memory Study. Neurology. 2013;81:896–903. 10.1212/WNL.0b013e3182a35228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Forette F, Seux ML, Staessen JA, Thijs L, Babarskiene MR, Babeanu S, et al. The prevention of dementia with antihypertensive treatment: new evidence from the systolic hypertension in Europe (Syst-Eur) study. Arch Intern Med. 2002;162:2046–2052. [DOI] [PubMed] [Google Scholar]
- 39. Shepherd J, Blauw GJ, Murphy MB, Bollen EL, Buckley BM, Cobbe SM, et al. Pravastatin in elderly individuals at risk of vascular disease (PROSPER): a randomised controlled trial. Lancet. 2002;360:1623–1630. [DOI] [PubMed] [Google Scholar]
- 40. Graves AB, van Duijn CM, Chandra V, Fratiglioni L, Heyman A, Jorm AF, et al. Alcohol and tobacco consumption as risk factors for Alzheimer’s disease: a collaborative re-analysis of case-control studies. EURODEM Risk Factors Research Group. Int J Epidemiol. 1991;20(Suppl 2):S48–S57. [DOI] [PubMed] [Google Scholar]
- 41. Salomon AR, Marcinowski KJ, Friedland RP, Zagorski MG. Nicotine inhibits amyloid formation by the beta-peptide. Biochemistry. 1996;35:13568–13578. [DOI] [PubMed] [Google Scholar]
- 42. Riggs JE. Smoking and Alzheimer’s disease: protective effect or differential survival bias? Lancet. 1993;342:793–794. [DOI] [PubMed] [Google Scholar]
- 43. Alkadhi KA. Chronic stress and Alzheimer’s disease-like pathogenesis in a rat model: prevention by nicotine. Curr Neuropharmacol. 2011;9:587–597. 10.2174/157015911798376307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Freathy RM, Kazeem GR, Morris RW, Johnson PC, Paternoster L, Ebrahim S, et al. Genetic variation at CHRNA5-CHRNA3-CHRNB4 interacts with smoking status to influence body mass index. Int J Epidemiol. 2011;40:1617–1628. 10.1093/ije/dyr077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lim SS, Vos T, Flaxman AD, Danaei G, Shibuya K, Adair-Rohani H, et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2224–2260. 10.1016/S0140-6736(12)61766-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Proitsi P, Lupton MK, Velayudhan L, Newhouse S, Fogh I, Tsolaki M, et al. Genetic predisposition to increased blood cholesterol and triglyceride lipid levels and risk of Alzheimer disease: a Mendelian randomization analysis. PLoS Med. 2014;11:e1001713 10.1371/journal.pmed.1001713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. [DOI] [PubMed] [Google Scholar]
- 48. Mannila MN, Mahdessian H, Franco-Cereceda A, Eggertsen G, de Faire U, Syvanen AC, et al. Identification of a functional apolipoprotein E promoter polymorphism regulating plasma apolipoprotein E concentration. Arterioscler Thromb Vasc Biol. 2013;33:1063–1069. 10.1161/ATVBAHA.112.300353 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(TIF)
n = 16,691.
(TIF)
(TIF)
SNPs from all scores (N unique = 302) were LD-pruned and duplicates were removed to leave the 269 variants shown here. The SNP near APOE (rs6857) had a p-value of 2.5 × 10−575, which was truncated to 10−30 for display on the figure.
(TIF)
SNPs from all scores (N unique = 302) were LD-pruned and duplicates were removed to leave 269 variants. For the present plot, the SNP near APOE (rs6857, p = 2.5 × 10−575) was excluded (see S4 Fig).
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
*Note that the effect sizes for the effects of SNPs on smoking quantity were estimated in current smokers only, while the associations with AD are not stratified by smoking status.
(TIF)
SNPs included in each risk score, their associations estimated by the relevant consortium, and their association with AD (IGAP [12]). Proxies are marked by an asterix in the leftmost column. The effect allele frequency was based on data from the EPIC-InterAct study [26]. Double asterisks indicate that frequency information was not available in the EPIC-InterAct study, and were obtained from European 1000 Genomes samples.
(XLSX)
(DOCX)
Data Availability Statement
All association data with Alzheimer's disease for each individual SNP is available from http://www.pasteur-lille.fr/en/recherche/u744/igap/igap_download.php. Individual-level data can be sought by request to the GERAD consortium (http://gtr.rcuk.ac.uk/project/B6C58A7C-3C3E-41CB-AF10-16DB59962C9E). To promote collaboration and wide use of data in order to further Alzheimer’s Disease research, qualified investigators can submit proposals to conduct analyses using results from the Genetic and Environmental Risk in Alzheimer’s Disease (GERAD) analysis or, in collaboration with GERAD, to conduct analyses using data from GERAD members. The PI and other key personnel must be qualified investigators with appropriate experience and have the support of at least one GERAD collaborator. Proposals can be submitted at any time and will be provided to all GERAD members, who will be asked to review and feedback to the GERAD coordinator (Julie Williams) within 2 weeks. Where objections or additional information is sought the proposal may be discussed on the next GERAD conference call. Further revision or clarification may be requested at this time. After all questions are addressed, the GERAD consortium will provide any objections to the coordinator (Julie Williams). All objections will be taken into account and acted on appropriately. Individual members can opt-out of projects. Proposals should not overlap with analyses that are being performed within GERAD, and should be new and novel research ideas. In order to access GERAD data, the receiving Institution is required to enter into a data sharing agreement with Cardiff University which binds them to appropriate use and acknowledgement of the data they receive. Only parties involved in this agreement can access GERAD data for the purpose specified in the proposal.