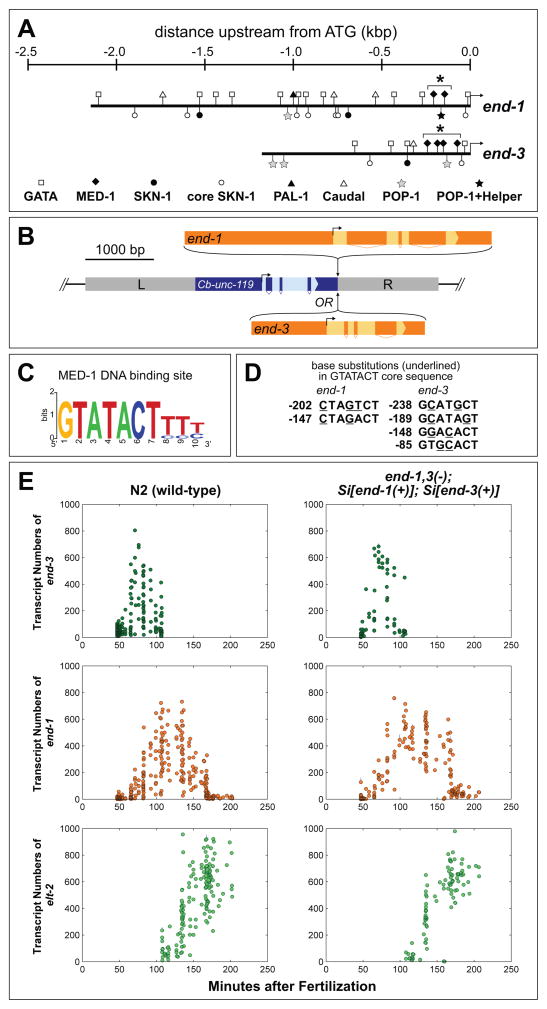
Fig. 2.
Construction of wild-type and MED-site mutated end-1 and end-3 single-copy transgenes. (A) Promoter structure of the end-1 and end-3 genes. The complete 5′ region up to the coding region of the nearest upstream gene is taken to be the 5′ end of the promoters. Putative binding sites for GATA factors (presumptive sites for END-1 and/or END-3), MED-1, SKN-1 and POP-1 are shown as described in Table S1. The cluster of MED-1 binding sites in each gene is indicated by an asterisk (*). The proximal POP-1 site in end-1 is adjacent to Helper sequences (Bhambhani et al., 2014). (B) General structure of MosSCI-mediated targeting constructs. Either end-1 or end-3 were inserted downstream of Cb-unc-119(+) in a vector to target the Mos insertion site in ttTi5605 II (for end-1) or ttTi4348 I (for end-3); homology arms (L and R) were appropriate to the insertion site. These insertion sites have been previously found to be compatible with normal expression of other transgenes, including those that are maternally expressed (Frokjaer-Jensen et al., 2012). (C) Structure of the MED-1 binding sites (Lowry et al., 2009). (D) Mutations introduced at the MED binding sites in end-1 or end-3. At least two base pairs were mutated within the GTATACT invariant core. The location of each site is given in bp relative to the ATG start codon. The mutations do not alter any other known binding sites, including those of the POP-1 Helper sequences (Bhambhani et al., 2014) or of a putative TATA box, as the MED site differs from the TATA consensus (Grishkevich et al., 2011; Lowry et al., 2009). (E) Expression levels in transcripts per embryo of end-3, end-1 and elt-2. Each dot represents a single embryo. Lines across each dot represent a 95% confidence interval for the expression level of that embryo. Minutes after fertilization is based on mapping to nuclei count to time at 25°C using data from a prior report (Bao et al., 2006).