Figure 1.
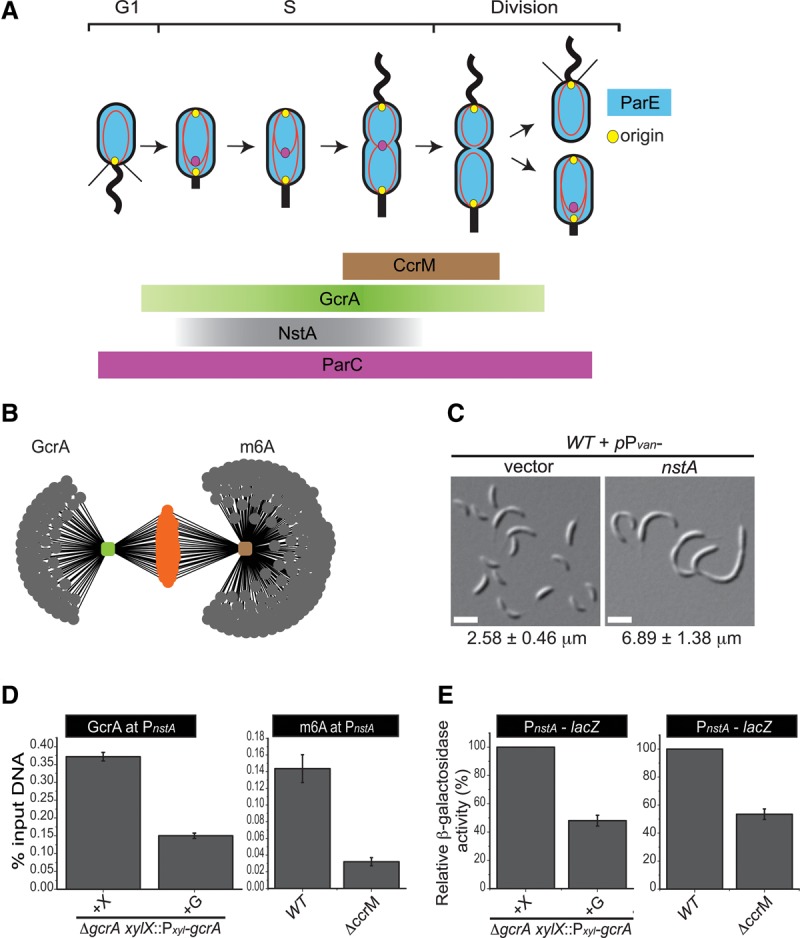
S-phase-specific developmental regulators in C. crescentus. (A) Schematic showing the cell cycle abundance of the S-phase-specific regulatory molecules CcrM (brown) and GcrA (green) and the topo IV regulator NstA (negative switch for topo IV decatenation activity) (gray). Also shown are the replication and sequestration of the newly replicated origin (yellow) and the presence of ParC (magenta) and ParE (blue) during the cell cycle in C. crescentus. (B) Network image showing putative promoters (gray dots) regulated by CcrM, detected using m6A (brown) and GcrA (green), as inferred from ChIP-seq (chromatin immunoprecipitation [ChIP] combined with “deep” sequencing) results of wild-type C. crescentus (Fioravanti et al. 2013). The common targets between m6A and GcrA are shown in orange. (C) Differential interference contrast (DIC) microscopy images of wild-type (WT) C. crescentus cells harboring the high copy vector (pMT335) or overexpressing nstA from the vanillate-inducible promoter Pvan on pMT335. Vanillate (0.5 mM) induction was done for 6 h. Mean cell size ± SD of at least 200 cells is given at the bottom of the image. Bar, 2 μm. (D) Data of qChIP (quantitative ChIP) experiments showing the occupancy of GcrA and m6A at PnstA in gcrA-depleted or ccrM-deleted cells. (E) Relative percentage of β-galactosidase activity of the PnstA-lacZ reporter in wild-type, gcrA-depleted, and ccrM-deleted cells. The gcrA xylX::Pxyl-gcrA cells were grown in the presence of xylose (+X) or glucose (+G) for 9 h. The values, ±SE, denoted in D and E are the average of three independent experiments.
