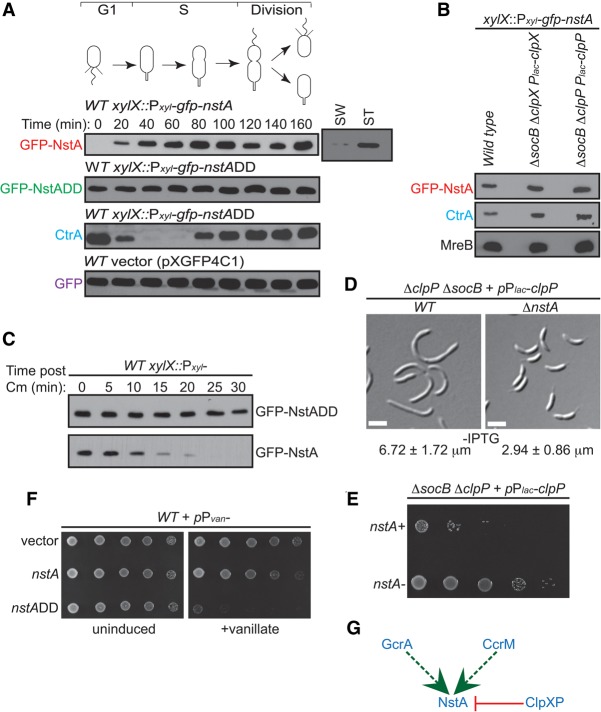
Figure 2.
Regulation of NstA abundance during the cell cycle by the ClpXP protease. (A) Immunoblot analyses in synchronized populations of C. crescentus to determine the relative abundance of GFP-NstA, GFP-NstADD, CtrA, and GFP during the cell cycle. Wild-type (WT) cells harboring the plasmids for gfp-nstA, gfp-nstADD, and gfp at the xylX locus on the chromosome were used. Cells were treated with 0.3% xylose. Levels of GFP-NstA in swarmer (SW) and stalked (ST) cell populations of resynchronized cells at 140 min are also shown. (B) Immunoblots of GFP-NstA, CtrA, and MreB in wild-type cells and in ΔsocB cells depleted for clpX or clpP. The strains were not treated with IPTG, the inducer for the Plac promoter. MreB was used as the loading control. (C) Immunoblots showing the difference in stability of GFP-NstA and GFP-NstADD. Expression of gfp-nstA or gfp-nstADD was induced with 0.3% xylose for 4 h prior to the inhibition of translation by chloramphenicol (Cm) treatment. The abundance of GFP-NstA or GFP-NstADD was monitored over time as indicated. (D) DIC microscopy images of ΔsocB cells depleted for clpP in the presence and absence of nstA. Cells were not treated with IPTG. Mean cell size ± SD of at least 200 cells is given at the bottom of the image. Bar, 2 μm. (E) Dilution plate showing the growth of cells from D. Fivefold serial dilutions of cells from D were spotted onto medium without IPTG. (F) Growth of wild-type strains harboring the high copy vector (pMT335) and expressing nstA or nstADD from the Pvan promoter on pMT335. Fivefold dilutions of the indicated strains were spotted onto medium with or without the inducer vanillate (0.5 mM). (G) Schematic summarizing the transcriptional and post-translational regulation of NstA. (Dashed green line) Positive transcriptional regulation by GcrA and CcrM; (red line) negative post-translational regulation by the protease ClpXP.