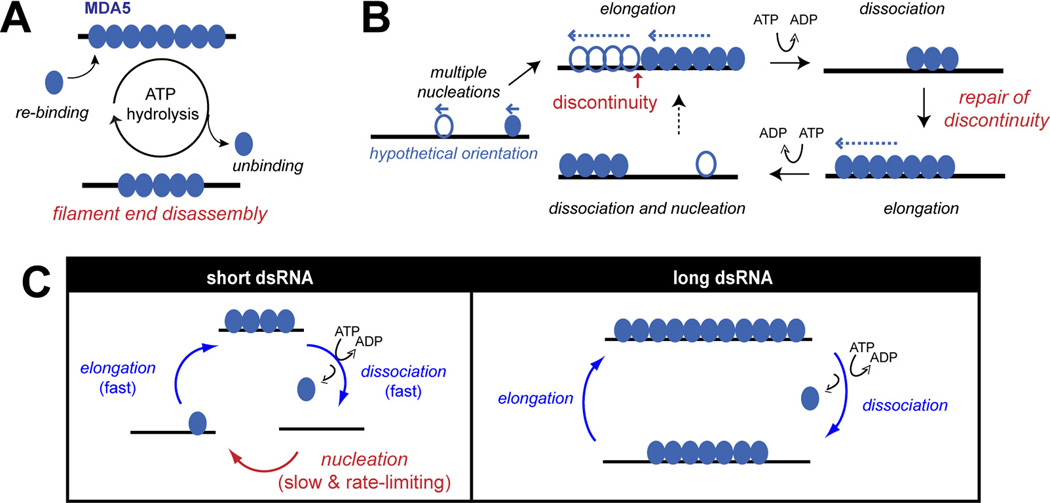
Fig 3.
Kinetic model for dsRNA length discrimination by the MDA5 filament. (A) ATP hydrolysis promotes end-disassembly of the MDA5 filament, which results in dsRNA length-dependent stability. (B) Multi-nucleated filament repairs gaps (or discontinuity) during ATP-mediated filament rearrangement. During dynamic equilibrium between assembly and ATP-driven end disassembly reactions, short filaments disassemble faster than longer ones, allowing continued elongation of longer filaments to more accurately reflect the length of underlying dsRNA scaffolds. (C) The role of slow nucleation kinetics in dsRNA length discrimination by MDA5. On short dsRNA, RNA binding occurs primarily by rate-limiting nucleation, whereas on long dsRNA, it is mediated by rapid filament elongation. Thus, slow nucleation selectively suppresses MDA5 binding to short dsRNA.