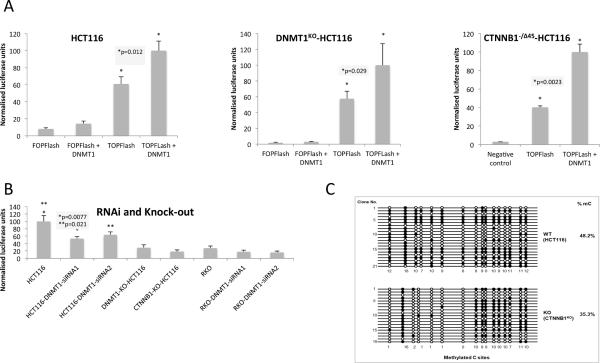
Figure 7. Functional consequences of the Dnmt1- β-catenin interaction.
(A) TCF (luciferase) reporter activity in HCT116 and CTNNB1−/Δ45-HCT116 (mutant β-catenin) cells(B) TCF (luciferase) reporter activity in HCT116 and RKO cells (treated with DNMT1 siRNAs) as well as DNMT1KO-HCT116 and CTNNB1KO-HCT116 cells. All cells were transfected with LEF/TCF reporter. Negative and positive controls were also used for signal normalization and transfection efficiency monitoring. Dual Luciferase assay was performed 48 hours after transfection and promoter activity values were expressed as arbitrary units using a Renilla reporter for internal normalization. Experiments were done in triplicates for biological cell cultures/ transfections and luminescent measurements. Average numbers (bar height) of relative luciferase units for each sample plus standard deviation (error bar) were plotted relative to maximum. (C) Methylated CpG sites are decreased in CTNNB1 KO cells at the H19 locus, especially on the first six (1, 3, 4, 5, 6, 7) of seven CpG sites. Genomic DNAs from wild-type HCT116 (WT) and CTNNB1 knockout HCT116 (KO) clones were bisulfite treated. The CpG island of H19 locus was PCR amplified, cloned, and sequenced with the Sanger sequencing method. Twenty one and nineteen clones were sequenced for WT and KO, respectively (methylated CpG sites are black spots, non-methylated CpG sites are white spots and DNA sequences are represented as black lines).