Fig. 1.
Kinase activity and inhibition of FGFR1 variants.
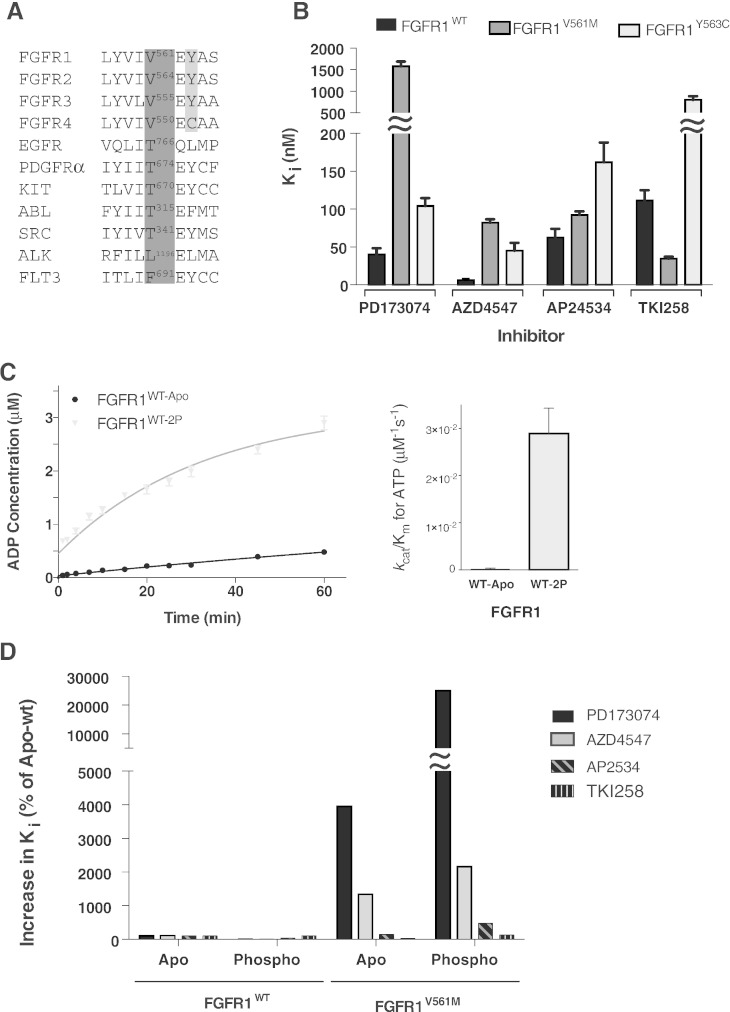
A. Amino acid alignment of part of the hinge region of the FGFR family and a number of other tyrosine kinases with reported gatekeeper mutations. The gatekeeper residue in each case is numbered and bordered in dark grey. The FGFR 1 to 3 tyrosine that is a cysteine in FGFR4 is highlighted in light grey.
B. Histogram showing the inhibition constants (Ki) for four selected compounds upon FGFR1 kinase domain protein, wild type and the indicated mutants. The data were generated from enzyme kinetic analyses using various concentrations of inhibitors and fitting using the Morrison equation within Graphpad Prism. Each data point was repeated in duplicate and the standard error of the mean is presented on each bar.
C. Left panel: comparison of the activity of FGFR1 apo (WT-apo) and phosphorylated (WT-2P) kinase domain protein produced using the ADP-Glo Assay. Each data point was produced in triplicate and the standard errors are indicated. Right panel: comparison of the enzyme efficiency (kcat/Km) of FGFR1 apo and phosphorylated protein. Parameters were generated through Michaelis–Menten kinetic experiments and analysed using Graphpad Prism software.
D. Histogram showing the change in inhibition constants for phosphorylated FGFRWT and apo and phosphorylated FGFR1V561M when normalised to apo FGFR1WT values (taken as 100%) for the indicated inhibitors.
The data in B–D are representative for 3–4 independent experiments.
See also Supplemental Tables S1 and S2.