Figure 1.
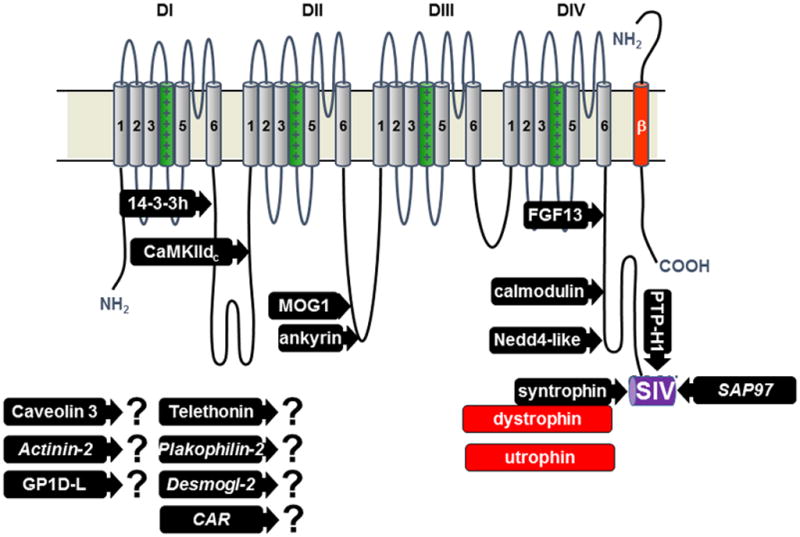
Topography of NaV1.5 channel and its interacting proteins. The proteins for which a binding site has been mapped are represented: 14-3-3 protein _-isoform, calmodulin-dependent protein kinase II delta-c, MOG1, ankyrin-g, fibroblast growth factor like 13, calmodulin, Nedd4-2 like ubiquitin ligases, syntrophin proteins adapting either dystrophin or utrophin, protein tyrosine phosphatase-H1, synapse associated protein-97. The proteins with question marks were found to interact with NaV1.5 but the sites of interaction are not yet known (CAR is coxsackie and adenovirus receptor, Desmogl-2 is desmoglein-2). Only one of the four beta subunits is represented (in red).
