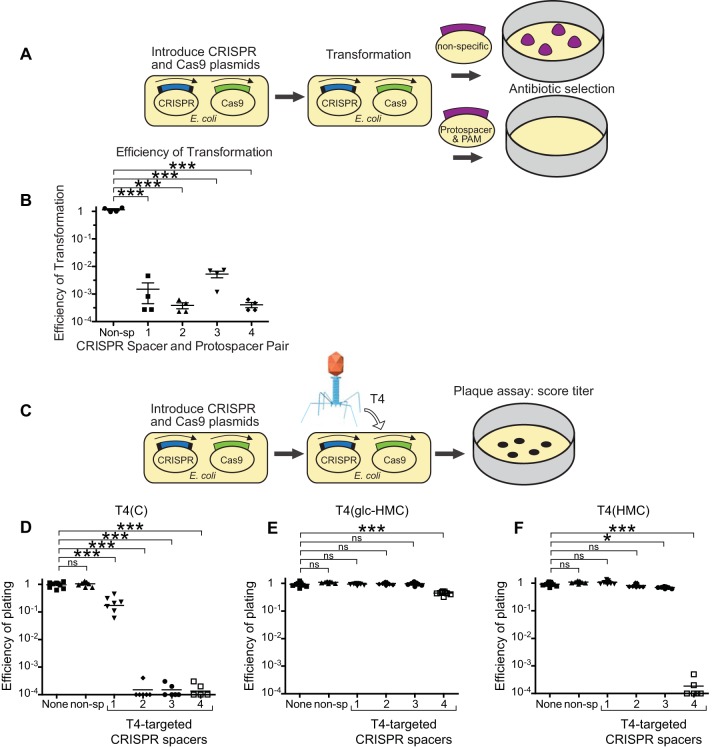
FIG 4 .
Glc-HMC and HMC modifications inhibit attack by the CRISPR-Cas9 system on phage T4. (A) Diagram of the strategy used to validate CRISPR spacers in a transformation assay. Bacteria containing the type II CRISPR system were transformed with a pUC19 plasmid containing either a T4 protospacer and PAM sequence or a nonspecific DNA sequence. Antibiotic selection for the pUC19 plasmid and quantification of the efficiency of transformation reveal the efficacy of CRISPR system cleavage of unmodified DNA containing a protospacer and PAM. (B) Results of plasmid challenge tests. The efficiency of transformation is the ratio of colony counts of cells transformed with equal amounts of pUC19 that contain a protospacer targeting the plasmid (numerator) to the colony counts of cells transformed with pUC19 (denominator). (C) Diagram of plaque assays to assess inhibition of T4 infection with CRISPR-Cas9. (D to F) Results of plaque assays in which the E. coli strains indicated were infected with up to 1 × 104 PFU of T4(C) (panel D), T4(glc-HMC) (panel E), or T4(HMC) (panel F). E. coli strains expressed Cas9 and crRNAs targeting T4 or controls. Starting from the left in each panel, None indicates no crRNA or Cas9, non-sp indicates nonspecific crRNA, 1 contained the maximum number of cytosines in the target strand and seed sequence, 2 contained the maximum number of cytosines in the target and complementary strands, 3 contained no cytosines in the target strand and seven cytosines in the complementary strand, and 4 contained the fewest cytosines in the target and complementary strands. Mean values were compared with the Kruskal-Wallis test. *, P < 0.01; ***, P < 0.0001; ns, not significant.