Figure 5.
T-CAD Is Expressed by Both Neurons and BVs at Peak Differentiation and Co-alignment
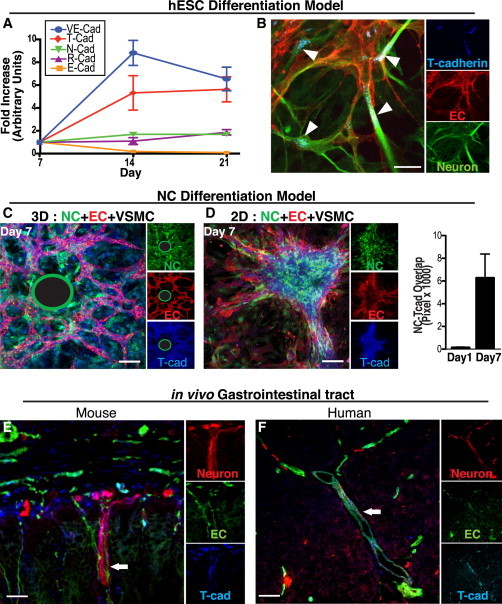
(A) Q-PCR analysis of cadherin gene expression in the hESC differentiation model. T-CAD (red diamonds) expression is upregulated by day 14, when co-patterning is observed and when VE-cadherin (an EC marker) expression is highest (n = 2 replicates, two independent experiments).
(B) T-cad (blue) staining in the hESC differentiation model. T-cad is expressed on both ECs (U.E. lectin, red) and neurons (βIII-tubulin, green) at day 21. Arrowheads denote T-cad localized to EC/neuronal junctions. Scale bars represent 25 μm.
(C and D) T-cad (blue) is expressed in NCs (green) only when cultured with both ECs (U.E. lectin, red) and VSMCs in our NC differentiation model. This was observed in 3D (C) and 2D (D). (The green-encircled black regions represent the location of the Cytodex beads upon which ECs were coated in the 3D fibrin gel system; see Supplemental Experimental Procedures).
(D, right) Quantitation of NC (green) co-localization with T-CAD staining (blue) (based on overlap of pixels) increasing over time (n = 5 images, two independent experiments). Data are presented as mean ± SEM. Scale bar represents 100 μm.
(E) Immunostaining of mouse GI tract for T-cad (blue) on ECs (CD31, red) and neurons (L1, green).
(F) Immunostaining of human GI tract for T-cad (blue) on ECs (U.E. lectin, red) and neurons (βIII tubulin, green). White arrow in both the mouse (E) and human (F) gut indicates an example of the typical close juxtaposition of NC-derived enteric neurons and vasculature seen with T-cad expression interposed between the two lineages. Scale bar represents 25 μm. See Figure S5 to observe the loss of neurovascular co-patterning in the gut of the T-cad KO mouse with consequent reduction of NC-derived neurons leading to a condition known clinically as a Hirschsprung’s (reduced ganglionic) phenotype. In (C)–(F), the large panel is a merged image of the three markers shown individually as insets at the right of each figure.
