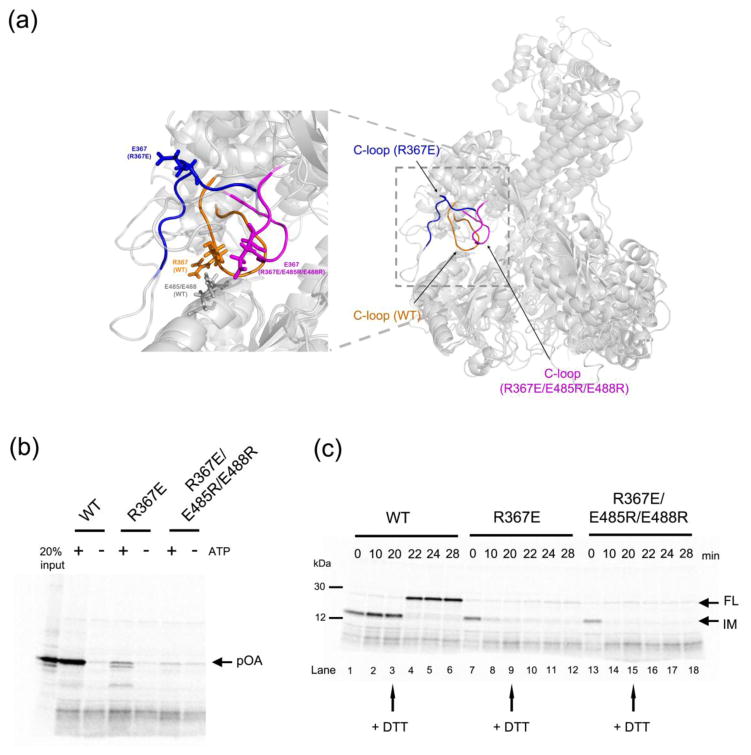
Figure 4. Analysis of SecA mutants defective in clamp opening or closure.
(a) Comparison of the molecular dynamics simulations of wild type SecA (C-loop residues 360-370 in orange), a SecA mutant (R367E) defective in maintaining a closed clamp conformation (C-loop in blue), and a SecA mutant defective in clamp opening (R367E/E485R/E488R; C-loop in magenta). The alignment is based on NBD2. The left panel shows a magnified view of the interaction between the C-loop and NBD2, with residue 367 in all SecA proteins and E485/E488 in wild type SecA shown in stick presentation.
(b) Translocation assays were performed by incubating 35S-methionine labeled proOmpA (pOA) with wild type or mutant SecA and proteoliposomes containing SecY complex. The reactions were performed in the presence or absence of ATP. Proteinase K was added and material protected from proteolysis was analyzed by SDS-PAGE and autoradiography. The left most lane shows 20% of the input material.
(c) A translocation intermediate was generated with proOmpA containing a disulfide bridge at its the C-terminus in the presence of His-tagged SecA and proteoliposomes containing SecY complex and Ni-NTA lipids. The samples were incubated with imidazole and either wild type or mutant SecA, followed by addition of DTT as indicated by arrows. Samples taken at the indicated time points were treated with proteinase K and analyzed by SDS-PAGE and autoradiography. IM, intermediate; FL, full-length protein.