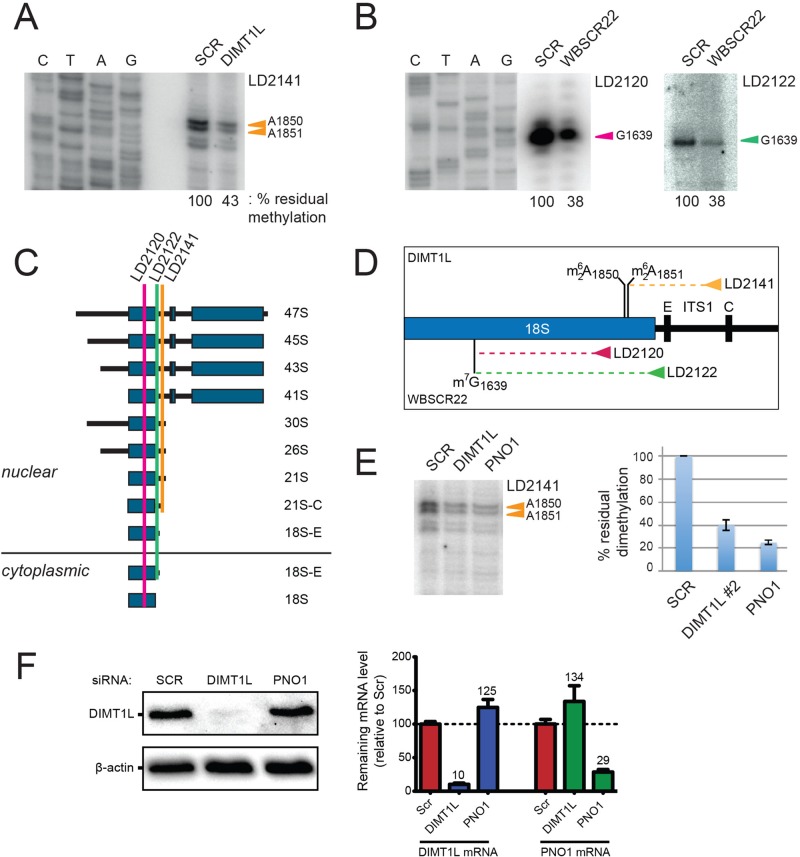
FIGURE 6:
DIMT1L and WBSCR22 are required for 18S rRNA 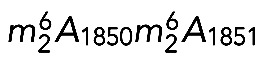 and m7G1639 methylation, respectively. (A–D) Primer extension mapping of the substrate nucleotides of DIMT1L and WBSCR222 on human 18S rRNA. Total RNA was extracted from HeLa cells depleted for 3 d with a DIMT1L-targeting (A) or WBSCR22-targeting (B) siRNA and analyzed by primer extension with oligonucleotide LD2120, LD2122, or LD2141. (C, D) The positions of the oligonucleotides and of the rRNA species. (E) PNO1 is required for efficient DIMT1L-mediated dimethylation. Total RNA was extracted from HeLa cells depleted for 3 d with a siRNA specific to DIMT1L or PNO1 and processed by primer extension with oligonucleotide LD2141. The level of residual dimethylation estimated with a Phosphorimager is indicated to the right. The results shown are means of three independent experiments. The sequences of the siRNAs used are listed in Supplemental Table S3 (WBSCR22#1, DIMT1L#2). (F) PNO1 is not required for the metabolic stability of DIMT1L. Total protein extract from HeLa cells depleted for 3 d with a siRNA specific to DIMT1L or PNO1 and tested by Western blotting with the antibodies indicated. Right, residual level of DIMT1L and PNO1 mRNAs assessed by qRT-PCR.
and m7G1639 methylation, respectively. (A–D) Primer extension mapping of the substrate nucleotides of DIMT1L and WBSCR222 on human 18S rRNA. Total RNA was extracted from HeLa cells depleted for 3 d with a DIMT1L-targeting (A) or WBSCR22-targeting (B) siRNA and analyzed by primer extension with oligonucleotide LD2120, LD2122, or LD2141. (C, D) The positions of the oligonucleotides and of the rRNA species. (E) PNO1 is required for efficient DIMT1L-mediated dimethylation. Total RNA was extracted from HeLa cells depleted for 3 d with a siRNA specific to DIMT1L or PNO1 and processed by primer extension with oligonucleotide LD2141. The level of residual dimethylation estimated with a Phosphorimager is indicated to the right. The results shown are means of three independent experiments. The sequences of the siRNAs used are listed in Supplemental Table S3 (WBSCR22#1, DIMT1L#2). (F) PNO1 is not required for the metabolic stability of DIMT1L. Total protein extract from HeLa cells depleted for 3 d with a siRNA specific to DIMT1L or PNO1 and tested by Western blotting with the antibodies indicated. Right, residual level of DIMT1L and PNO1 mRNAs assessed by qRT-PCR.