Figure 2.
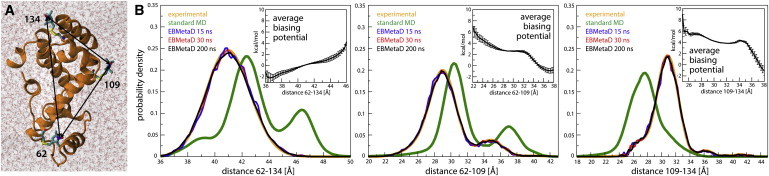
(A) Spin-labeled T4 lysozyme simulated in explicit water (PDB:2LZM (13)). The protein is enclosed in a truncated-octahedron periodic box containing 11,895 TIP3P water molecules and 10 Cl− counterions that neutralize the total charge of the system. The distances between the spin-label nitroxide groups measured by ESR/DEER are indicated (solid arrows). (B) Comparison of the experimental and calculated probability distributions for each of the spin-label pairs, from either unbiased MD simulations or EBMetaD; the latter are given for different simulation times. (Insets) EBMetaD biasing potential, averaged over the simulated trajectory (te = 5 ns). Error bars are standard errors over three simulation fragments (see Fig. S4 for further details).
