Abstract
Copy number variation (CNV) is a source of genetic diversity in humans. Numerous CNVs are being identified with various genome analysis platforms, including array comparative genomic hybridization (aCGH), single nucleotide polymorphism (SNP) genotyping platforms, and next-generation sequencing. CNV formation occurs by both recombination-based and replication-based mechanisms and de novo locus-specific mutation rates appear much higher for CNVs than for SNPs. By various molecular mechanisms, including gene dosage, gene disruption, gene fusion, position effects, etc., CNVs can cause Mendelian or sporadic traits, or be associated with complex diseases. However, CNV can also represent benign polymorphic variants. CNVs, especially gene duplication and exon shuffling, can be a predominant mechanism driving gene and genome evolution.
Keywords: FoSTeS, genomic disorder, genomotype/phenotype correlations, MMBIR, NAHR, NHEJ
INTRODUCTION
The tremendous variability of human genomes highlights the almost absurd notion of a single reference human genome sequence. Watson-Crick base-pair changes have long been well known; those with frequency >1% are referred to as single nucleotide polymorphisms (SNPs). Millions of SNPs have been revealed in human populations, and many more are being discovered with the determination of each personal genome sequence (Table 1) (10, 81, 159, 174, 177). Although numerous rare point mutations are known to cause sporadic disease, and SNPs are associated with common human diseases (http://www.genome.gov/gwastudies/), few gene rearrangements had been associated with disease traits until relatively recently. For example, it was not until the early 1990s that relatively large (>1 Mb) but submicroscopic genomic duplications and deletions causing gene CNV (copy number variation) were shown to cause Mendelian traits (16, 97, 99). CNVs like SNPs can also represent benign polymorphic variants present in >1% of the population. However, the extent to which large duplication and deletion CNVs contribute to human genetic diversity, and may or may not convey phenotypes, is still being unraveled.
Table 1.
Copy number variation (CNV) versus single nucleotide polymorphism (SNP)
| CNV (Database of Genomic Variants, http://projects.tcag.ca/variation/) |
SNP (dbSNP, http://www.ncbi.nlm.nih.gov/SNP/) |
|
|---|---|---|
| Total number | 38,406a (Mar 11, 2009) | 14,708,752 (Build 129) |
| Size | 100 bp to 3 Mb | Mostly 1 bp |
| Type | Deletion, duplication, complex | Transition, transversion, short deletion, short insertion |
| Effects on genes | Gene dosage, interruption, etc. | Missense, nonsense, frameshift, splice site |
| Percentage of the reference genome covered |
29.74%b | <1% |
including inversions.
This value may be overestimated owing to the resolution limitation of the technologies (e.g., BAC aCGH) used in CNV screening but also could be underestimated because of the inability to resolve smaller CNVs (1- to 20-kb range) and the limitations of the current reference genome.
In 2004, with the advent of genome-wide analysis tools that could be used to interrogate DNA content, two studies revealed that copy number variations (CNVs) [i.e., DNA segments that present at variable copy number in comparison to a reference genome with the usual copy number of N = 2 (35)] are widespread in human genomes and represent a significant source of genetic variation (57, 138). With the aid of genome-wide technologies of higher resolution, over 38,000 CNVs (>100 bp in size) and many other structural variations (SVs, including balanced inversions and translocations) have been reported (Table 1). In terms of total bases involved, SVs may account for more differences among individuals than do SNPs (132). In addition, CNVs appear to have a much higher de novo locus-specific mutation rate than SNPs based on estimations from CNV-associated disease prevalence (92) and observations from PCR assays of pooled sperm DNA (166).
How has the human genome come to have such a great amount of CNV? Two recombination-based mechanisms, i.e., nonallelic homologous recombination (NAHR) and nonhomologous end-joining (NHEJ) (98), and retrotransposition (63, 64, 68, 181), have been implicated in genomic rearrangements and the formation of CNVs. Recently, a novel replication-based mechanism, fork stalling and template switching (FoSTeS), has been proposed to account for the observed complex genomic rearrangements that cannot be readily explained by NAHR, NHEJ, or retrotransposition (78). Notably, breakpoint sequencing data also suggest that a portion of CNV likely occurs by a mechanism consistent with FoSTeS (125).
In addition to their association with sporadic and Mendelian diseases in humans, CNVs have also recently been shown to be associated with human complex traits such as susceptibility to HIV infection, autism, and schizophrenia (Table 2). Since CNV can encompass part or all of a gene, or be a genomic segment containing several genes, some CNVs are likely to have a role in the alteration of human physiological functions, Thus, as well as causing disease, human-specific CNVs may be responsible for the emergence of advantageous human-specific traits, such as cognition and endurance running (24, 91). They therefore are likely to be subject to evolutionary pressures such as selection as well as genetic drift (119).
Table 2.
Examples of copy number variations (CNVs) and conveyed genomic disordersa
| Phenotype | OMIM | Locus | CNV | Referencesa |
|---|---|---|---|---|
| Mendelian (autosomal dominant) b | ||||
| Williams-Beuren syndrome | 194050 | 7q11.23 | del | S4 |
| 7q11.23 duplication syndrome | 609757 | 7q11.23 | dup | S62 |
| Spinocerebellar ataxia type 20 | 608687 | 11q12 | dup | S30 |
| Smith-Magenis syndrome | 182290 | 17p11.2/RAI1 | del | S13 |
| Potocki-Lupski syndrome | 610883 | 17p11.2 | dup | S49 |
| HNPP | 162500 | 17p12/PMP22 | del | S11 |
| CMT1A | 118220 | 17p12/PMP22 | dup | S41 |
| Miller-Dieker lissencephaly syndrome | 247200 | 17p13.3/LIS1 | del | S10, S50 |
| Mental retardation | 601545 | 17p13.3/LIS1 | dup | S6 |
| DGS/VCFS | 188400/192430 | 22q11.2/TBX1 | del | S16, S55 |
| Microduplication 22q11.2 | 608363 | 22q11.2 | dup | S17, S47, S75 |
| Adult-onset leukodystrophy | 169500 | LMNB1 | dup | S48 |
| Mendelian (autosomal recessive) | ||||
| Familial juvenile nephronophthisis | 256100 | 2q13/NPHP1 | del | S31, S53 |
| Gaucher disease | 230800 | 1q21/GBA | del | S5 |
| Pituitary dwarfism | 262400 | 17q24/GH1 | del | S9, S24 |
| Spinal muscular atrophy | 253300 | 5q13/SMN1 | del | S43, S51 |
| beta-thalassemia | 141900 | 11p15/beta-globin | del | S29 |
| alpha-thalassemia | 141750 | 16p13.3/HBA | del | S25 |
| Mendelian (X-linked) | ||||
| Hemophilia A | 306700 | F8 | inv/del | S2 |
| Hunter syndrome | 309900 | IDS | del/inv | S8, S70, S72 |
| Ichthyosis | 308100 | STS | del | S56 |
| Mental retardation | 300706 | HUWE1 | dup | S21 |
| Pelizaeus-Merzbacher disease | 312080 | PLP1 | del/dup/tri | S14, S28, S37, S38, S71 |
| Progressive neurological symptoms (MR+SZ) | 300260 | MECP2 | dup | S3, S15, S65 |
| Red-green color blindness | 303800 | opsin genes | del | S46 |
| Complex traits | ||||
| Alzheimer disease | 104300 | APP | dup | S52 |
| Autism | 612200 | 3q24 | inherited homozygous del | S45 |
| 611913 | 16p11.2 | del/dup | S34, S42, S54, S68 | |
| Crohn disease | 266600 | HBD-2 | copy number loss | S20 |
| 612278 | IRGM | del | S44 | |
| HIV susceptibility | 609423 | CCL3L1 | copy number loss | S23, S33 |
| Mental retardation | 612001 | 15q13.3 | del | S58 |
| 610443 | 17q21.31 | del | S32, S57, S59 | |
| 300534 | Xp11.22 | dup | S21 | |
| Pancreatitis | 167800 | PRSS1 | tri | S36 |
| Parkinson disease | 168600 | SNCA | dup/tri | S12, S19, S22, S27, S61 |
| Psoriasis | 177900 | DEFB | copy number gain | S26 |
| Schizophrenia | 612474 | 1q21.1 | del | S63, S64, S67 |
| 181500 | 15q11.2 | del | S63 | |
| 612001 | 15q13.3 | del | S63, S64 | |
| Systemic lupus erythematosus | 152700 | FCGR3B | copy number loss | S1, S18, S69 |
| 120810 | C4 | copy number loss | S74 | |
For the supplemental references, follow the Supplemental Material link from the Annual Reviews home page at http://www.annualreviews.org.
Also de novo sporadic.
COPY NUMBER VARIATION AND STRUCTURAL VARIATION: THE PHENOMENON
In 2004, two studies reported that CNVs of many large DNA genomic segments exist between normal human individuals. Sebat et al. (138) employed a technology termed ROMA (representational oligonucleotide microarray analysis) with 85,000 interrogating probes with an average spacing of 35 kb to study the large-scale (>100-kb) copy number differences between 20 normal individuals. In total, 221 copy number changes were detected at 76 CNV loci. Using a BAC (bacterial artificial chromosome) CGH array (127) with resolution of ~1 Mb, Iafrate et al. (57) investigated large-scale CNVs in 55 unrelated individuals and identified 255 clones with copy number gain or loss. Interestingly, 102 (41%) of these copy number changes are observed in more than one individual (57), which suggests that these variations are not rare but may represent polymorphic variations. Furthermore, examination of the genomic content of such CNVs revealed that these genomic regions are not “junk” but include many functional genes involved in regulation of cell growth and metabolism (57), thus implicating CNVs in human traits, disease, and evolution.
These two early studies provided evidence for a more expanded view of human genetic variation. However, owing to the limited subject sample size, assay resolution, and genome coverage, the newly discovered CNVs detected only a small fraction of the CNVs expected to exist in human genomes. In subsequent years, many additional studies using a multitude of different high-resolution genome analysis platforms have advanced our knowledge of CNVs, and a comprehensive CNV map is beginning to emerge.
SNP genotyping data have also been used to investigate deletion polymorphisms. By assuming that genotypes of SNPs included in CNVs will violate the rules of Mendelian transmission, as initially determined for the CMT1A duplication CNV (97, 101). Conrad et al. (18) examined the transmission patterns of SNP genotypes in 30 European-derived trios (i.e. mother, father, and child) from Utah and in 30 Yoruba African trios. A total of 586 deletion loci were identified, ranging in size from 300 bp to 1200 kb (18). Intriguingly, the size versus frequency distribution of these deletions follows an L-shaped curve; i.e., there are more small deletions and fewer large ones (18). A similar CNV distribution pattern was also found in other studies (Figure 1).
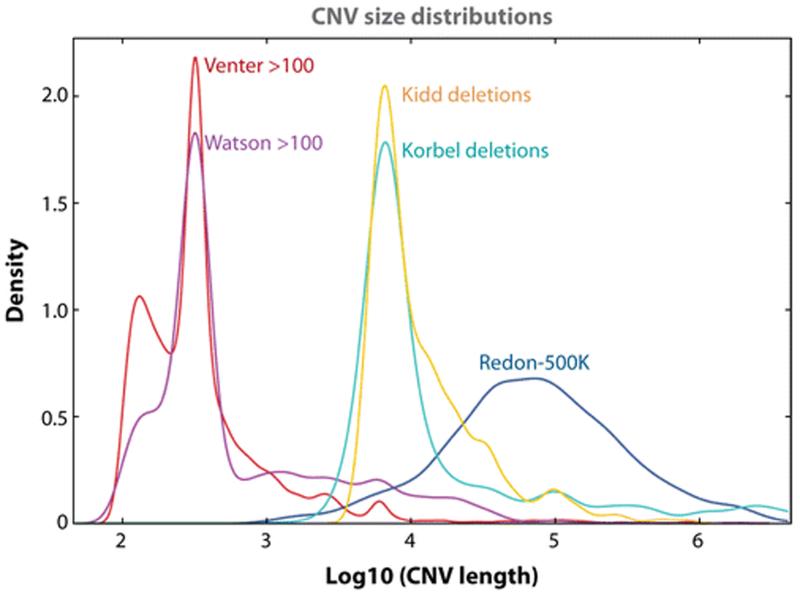
Figure 1.
Size distribution of copy number variations (CNVs) larger than 100 bp. Red, CNVs in the Venter genome (81); purple, CNVs in the Watson genome (177); blue, CNVs in Redon et al. (132); green, deletion CNVs in Korbel et al. (68); yellow, deletion CNVs in Kidd et al. (64). Note the greater detection of smaller-sized CNVs with higher-resolution genome analysis.
In a related study, McCarroll et al. (102) investigated the physically clustered patterns of null genotypes, apparent Mendelian inconsistencies and Hardy-Weinberg disequilibrium for the Phase I data (1.3 million SNPs, 269 individuals) of the International HapMap Project (159). They identified 541 deletion variants ranging from 1 kb to 745 kb in size, 278 of which are detected in multiple individuals. Hinds et al. (54) directly examined on resequencing arrays the reduced intensity of the hybridization signal caused by deletions and identified 215 relatively small deletions (70 bp to 10 kb) in 24 unrelated individuals. The deletion variations identified in these three studies added over 1000 CNVs (26) to a database where structural variation is cataloged (the Database of Genomic Variants or DGV, http://projects.tcag.ca/variation/).
In 2006, Redon et al. (132) constructed a first-generation CNV map of the human genome. They employed both SNP genotyping arrays (Affymetrix GeneChip Human Mapping 500K, 474642 SNPs) for comparative analysis of hybridization intensities and Whole Genome TilePath BAC arrays (WGTP, 26574 large insert BAC clones) for CGH in 270 HapMap individuals (159). A total of 1447 CNV regions were identified and approximately half of these were detected in multiple individuals (132).
The CNVs included in the DGV database are reported to account for 29.7% of the human genome (Table 1). However, in some of the initial studies, CNV size was often overestimated because of the relative low resolution of some platforms (e.g., BAC arrays) used in CNV screening. With the aid of high-resolution (~1 kb) aCGH, Perry et al. (125) studied 2191 known CNV regions in 30 individuals from 4 HapMap populations. They detected copy number changes in 1153 loci and narrowed the boundaries of 1020 (88%) CNV regions (125). Reduced CNV sizes were also reported in another study of McCarroll et al. (104) on the HapMap cohort via hybrid SNP-CNV genotyping arrays. Hence, the large-scale CNVs may affect less than previously proposed and CNVs perhaps encompass 5% of the human genome (104), although this estimate is still challenged by methods that do not resolve end points to the base-pair level and by arrays that are based on a reference genome that is limited in scope.
In addition to the array-based platforms (SNP or CGH arrays), CNV can also be investigated by DNA sequencing. By mining insertion and deletion polymorphisms from split capillary reads of DNA resequencing traces that were generated by shotgun sequencing the genomic DNA of 36 individuals, Mills et al. (110) discovered 415,436 nonredundant indels and CNVs, ranging from 1 to 9989 bp in size.
Balanced SVs such as inversions that cannot be identified by CGH are detectable by paired-end sequencing techniques. Eichler and colleagues (167) first compared fosmid (a phage cloning vector with DNA packaging limited to ~40 kb) DNA sequences from a library constructed from the genomic DNA of individual NA15510 and identified 297 potential SVs varying in size from 8 kb to 1.9 Mb. In a related study (64), Eichler and colleagues constructed new fosmid libraries from eight HapMap samples (four Yoruba Africans and four non-African individuals) and sequenced both ends (termed paired-end sequencing), of approximately one million clones per genome. Combined with the previous observations in the NA15510 fosmid library (167), they validated 1695 SVs across 9 diploid human genomes, including 747 deletions, 724 insertions, and 224 inversions. 50% of these were found in multiple libraries (64).
By using next-generation sequencing technology to derive sequence of paired ends of 3-kb DNA fragments and computational mapping of DNA reads onto the reference genome, Korbel et al. (68) developed a high-throughput method, paired-end mapping (PEM), to investigate SVs in two female individuals, one African (NA18505), the other putatively European (NA15510). This strategy can identify deletions, inversions, mated insertions, and unmated insertions of ~3 kb or larger, as well as simple insertions of 2 to 3 kb (68). In total, 1297 SVs, including 853 deletions, 322 insertions, and 122 inversions, were identified. Korbel et al. confirmed only 41% of the previously reported deletions and inversions of NA15510 mainly due to 62% coverage of NA15510; however, they also detected 407 new SVs in NA15510 (68, 167). By comparing the SVs identified in NA15510 with those in NA18505, these authors found that 45% SVs were shared between NA15510 and NA18505 (68).
Recently, comparison of the complete genomic sequence of Craig Venter (a de novo build of human diploid genome delineated with conventional Sanger dideoxy technology) (81), James D. Watson (177), an African (NA18507) (10), and Asian individual (174) (the latter three with next-generation sequencing following a comparison to the human reference genome) has revealed millions of SNPs, many short indels and large SVs. These studies showed conclusively what had long been suspected: there is a continuous size distribution of SVs in the human genome, with smaller variants being the most frequent at almost all scales (Figure 1).
Interestingly, the size distribution of deletions observed in the Watson and Venter genomes (81, 177) exhibits a marked enrichment in the size range of 300–350 bases owing to the known retrotransposition-based polymorphism of Alu elements, and a less marked enrichment at ~6 kb owing to the retrotransposition-based polymorphism of L1 elements (Figure 1). Array-based methods were used to identify large (e.g., >50 kb) CNVs in these genomes. For the Watson genome, the read depth of 454 sequencing confirmed many array-detected CNVs. However, many CNVs < 50 kb in size potentially remain to be identified. The paired-end sequencing methods used in the African and Asian genomes enabled smaller CNVs to be identified than was possible with array-based methods. However, due to the limit of read length (average 35 bases), large-scale CNVs (especially duplications) may have been missed by massively parallel sequencing (10, 174). Thus it remains a challenge to detect all CNVs in an individual genome using one single technology since CNV can range in size from single exons (~100 bp) to millions of base pairs. However, in principle it should be possible to use new sequencing technologies to identify all forms of SV by combining paired-read analyses with read-depth analyses and de novo assembly. Hence our current inability to capture all of these variants remains, in part, an analytical challenge rather than a technical one.
In addition to sequencing-based platforms, some inversions can also be studied by PCR-based approaches (39). NAHR between intrachromosomal low-copy repeat (LCR) sequences in reverse orientation can lead to inversion (154). Flores et al. (39) searched such potential recombinogenic inverted sequences (PRIS) in the human reference genome and identified a total of 24,547 PRIS, varying in size from 400 to 74868 nucleotides, which represent a huge resource of potential loci susceptible to inversion variation. Due to the PCR efficiency and technological limitations, these authors (39) selected eight PRIS for further study and identified inversion variations at six PRIS loci (75%), in two of which gene structures were affected by inversions.
To date (March 2009), the Database of Genomic Variants comprises 38,406 SVs (Table 1). Database limitations include the use of multiple platforms of varying degrees of genome resolution. Therefore, most CNVs are not resolved to the nucleotide level and have thus not been genotyped in different populations.
The contributions of CNVs, relative to SNPs, to human phenotypes, especially complex diseases, are still unknown. Recent studies attempted to partially explore the question: What are the relative contributions to phenotype of CNV and SNP when the phenotypic output measured is changes of gene expression? In a study of the genetic contribution to variation in gene expression of cell lines of 210 HapMap individuals, Stranger et al. (158) compared the SNP data of HapMap Phase I (160) to the CNV data of Redon et al. (132). After measuring the expression levels of 14,072 genes (14,925 transcripts), at least 8.75% to 17.7% of the variation in gene expression could be explained by CNVs (158) versus 92.5% to 83.6% that could be explained by SNPs. Remarkably few of the CNV associations were also observed at SNPs; i.e. there is minimal overlap between the contributions from CNV versus SNP. This could be potentially explained in part by the low power in this study due to the relatively small sample sizes in each HapMap population (158). In addition, because only a small portion of existing CNVs were investigated, the current observations in Stranger et al. (158) may underestimate the overall contribution of CNV to gene expression level. Further comprehensive research is expected to better elucidate the contributions of CNVs to human phenotypes.
How Many Copy Number Variations Remain to be Discovered?
Have we already identified all CNVs in the human genome? The answer is a definite “no”. First, the characterization of the SV in a certain genomic region requires knowledge about the sequence of this region. The current human genome sequence encompasses virtually all euchromatic regions; however, many gaps remain for which we have no sequence information. In the most recent draft published in April 2003 by the International Human Genome Sequence Consortium (59), up to 6% of the total genome is reported to remain uncovered, consisting of 341 sequencing gaps with 273 residing in euchromatin. An estimated 54% of all gaps in euchromatin regions are flanked by segmental duplications (SDs) or LCRs (5) and are thus prone to SVs (154). Many of these are adjacent to complex LCRs (LCRs consisting of a cluster of different repeat subunits lying in either orientation, see Figure 2 below) (154), such as the 15 gaps in the 1q21.1 deletion region recently found to be associated with microcephaly/macrocephaly and developmental/behavioral abnormalities (13, 106) and schizophrenia (Table 2). In aggregate, many of the current sequence gaps of the human genome can be expected to contain sequences with SV. In fact, SVs can be one of the factors that complicated the completion of sequencing in the gap regions because different haplotypes need to be differentiated from each other (14, 27). Most recently, Bovee et al. reported the closure of 26 of the 273 remaining gaps in euchromatin regions (12). Indeed, 30.7% of these closed gaps are polymorphic and show SVs (12).
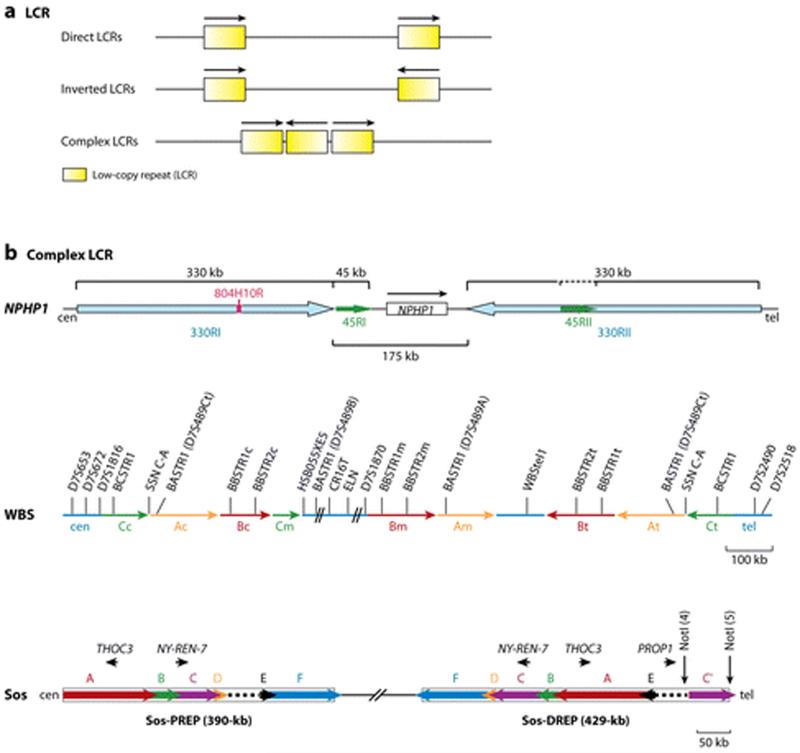
Figure 2.
Low-copy repeats (LCRs) or segmental duplications (SDs) in the human genome. (a) LCR orientations (direct, reverse, and complex LCRs). (b) Examples of complex LCR. NPHP1, the shaded blue representing the inverted 330-kb repeats and the green arrows for the direct 45-kb repeats (adapted from Reference 135). The WBS locus at 7q11.23, Blocks A, B, and C of centromeric (c), medial (m), and telomeric (t) LCRs represented by black arrows (adapted from Reference 9). The Sos locus, six subunit LCRs between the proximal and the distal Sos-REPs (A-F) and their orientation depicted by the arrowhead (adapted from Reference 72).
Even in the genomic regions with available reference sequence, we may identify more novel SVs in the future. Genomic structures such as the complex LCRs (Figure 2) containing multiplex adjacent LCR elements in tandem and reverse orientations provide the structural basis for a multitude of variations including duplication, deletion, inversion of different length and in diverse combinations (14, 20, 47, 154). The detection of all of these SVs will require detailed analyses of a large number of different haplotypes. Furthermore, complex LCRs, particularly those with inverted repeats, can also form cruciforms and may trigger genomic rearrangements, inducing nonrecurrent, sometimes complex SVs in some individuals. These variations tend to be rare and require large sample sizes for identification.
Finally, the reference sequence contains the deleted allele at a number of common CNVs, manifesting as novel inserted sequences in sequencing-based surveys (64), but missed by microarray-based surveys. These insertions can often be found in the chimp genome sequence, which suggests that these apparent insertions are indeed deletions in the human reference sequence.
Given the huge scientific and clinical significance of structural variations and human genomics in general, multicenter collaborative projects including the Human Genome Structural Variation Group (28, 64) and the 1000 Genomes Project (10, 174) have been launched to extend our knowledge on this subject. We are optimistic that these endeavors will unveil many novel structural variations of our genomes in the near future.
COPY NUMBER VARIATION: MECHANISMS FOR FORMATION
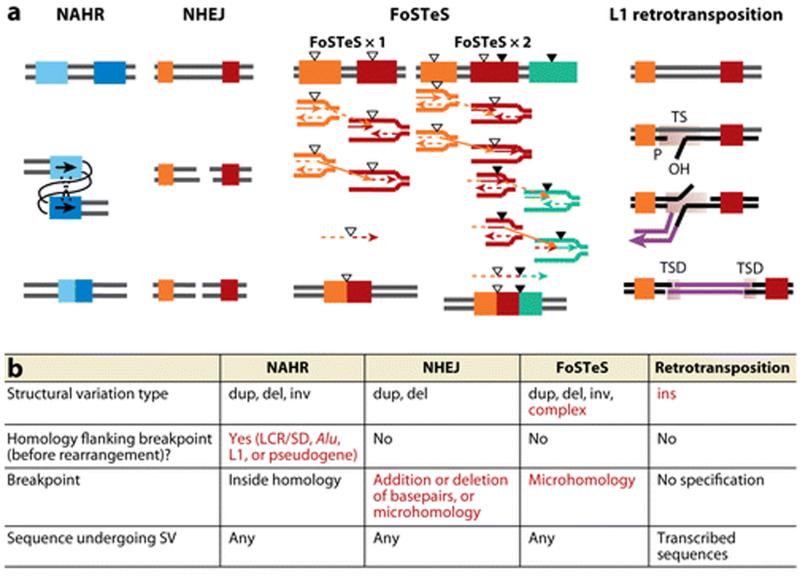
Four major mechanisms: NAHR, NHEJ, FoSTeS, and L1-mediated retrotransposition, generate rearrangements in the human genome and probably account for the majority of CNV (reviewed in References 47 and 51; Figure 3).
Figure 3.
Comparisons and characteristics of the four major mechanisms underlying human genomic rearrangements and CNV formation. (a) Models for Non-Allelic Homologous Recombination (NAHR) between repeat sequences (LCRs/SDs, Alu, or L1 elements); Non-Homologous End-Joining (NHEJ), recombination repair of double strand break; Fork Stalling and Template Switching (FoSTeS), multiple FoSTeS events (×2 or more) resulting in complex rearrangement and single FoSTeS event (×1) causing simple rearrangement; and retrotransposition. TS, target site; TSD, duplicated target site. Adapted from References (47, 123). Thick bars of different colors indicate different genomic fragments; completely different colors (as orange and red or orange/red/green in FoSTeS×2) symbolize that no homology between the two fragments is required. The two bars in two similar shades of blue indicate that the two fragments involved in NAHR should have extensive homology with each other. The triangles symbolize short sequences sharing microhomologies. Each group of triangles (either filled or empty) indicates one group of sequences sharing the same microhomology with each other. (b) Characteristic features for each rearrangement mechanism. Specific features of certain mechanisms are shown in red. Abbreviations: dup, duplication; del, deletion; inv, inversion; ins, insertion.
Nonallelic Homologous Recombination Occurs in Meiosis and Mitosis, Resulting in Duplication, Deletion, and Inversion
NAHR is caused by the alignment of and the subsequent crossover between two nonallelic (i.e., paralogous) DNA sequence repeats sharing high similarity to each other (Figure 3) (154). Repeats on the same chromosome and in direct orientation mediate duplication and/or deletion, whereas inverted repeats mediate inversion of the genomic interval flanked by the repeats (88, 154). NAHR between sequence repeats on different chromosomes can lead to chromosomal translocation (88, 154).
Substrates for NAHR are LCRs or SDs of >10 kb in length with 95%–97% similarity (Figure 2) (147, 154). Different groups of LCRs are sometimes localized adjacent to each other, with some subunits in tandem and others in reverse orientation resulting in complex LCRs (Figure 2) (15, 154). Complex LCRs can themselves undergo CNV in both meiosis and mitosis, and specific structural variants may predispose susceptibility to chromosomal rearrangements (7, 14, 20, 90). In addition to LCR, repetitive sequences such as the retrotransposable L1 elements (48), Alu (2, 49, 139), or matching pseudogenes (65, 157) can act as NAHR substrates if from similar families or with high enough sequence identity to facilitate homologous recombination. Such shorter sequence substrates tend to mediate NAHR of shorter DNA fragments (48, 139), which are often underestimated owing to the detection limit of many CNV assays (64, 68). A recombination hotspot of 28 bp has been detected in Alu sequence (139), whereas no recombination hotspots have been found in L1 sequences (48).
NAHR hotspots can be closely related to the allelic homologous recombination (AHR) hotspots (46, 60, 89, 163) and can overlap in the human genome and in our ancestral genomes (86, 114, 131). A cis-acting sequence motif CCNCCNTNNCCNC was found to be enriched in both AHR and NAHR hotspots (113) and may represent the analogue of Chi (GCTGGTGG) in prokaryotic homologous recombination (152).
NAHR can occur in meiosis, where it results in unequal crossing over as revealed by segregation of marker genotypes and leads to constitutional genomic rearrangements that can be benign polymorphisms or manifest sporadic, if de novo, or inherited genomic disorders (92, 98, 166). NAHR can also occur in mitosis, resulting in mosaic populations of somatic cells carrying copy number or SVs (39, 75, 76).
The recent use of the advanced array-based techniques enabled the identification of a number of novel genomic disorders caused by the predicted NAHR-mediated reciprocal genomic rearrangements (Table 2). The relative frequency of duplications and deletions mediated by the same pairs of LCRs is thus an increasingly important and clinically relevant question.
To address this question directly, Turner et al. (166) used pooled sperm PCR assays to analyze four NAHR loci related to well-studied genomic disorders, including Williams-Beuren syndrome (WBS; MIM 194050) deletion and 7q11.23 duplication, the azoospermia factor a (AZFa; MIM 415000) deletion and its reciprocal duplication, the hereditary neuropathy with liability to pressure palsies (HNPP; MIM 162500) deletion and Charcot-Marie-Tooth disease type 1A (CMT1A; MIM 118220) duplication, and the Smith-Magenis syndrome (SMS; MIM 182290) deletion and Potocki-Lupski syndrome (PTLS; MIM 610883) duplication in the sperm populations of five males. Strikingly, they found that all five subjects consistently displayed an approximately 2:1 ratio of deletion versus duplication at all three autosomal loci and 4:1 for the AZFa locus on the Y chromosome (166). Thus, at least in meiosis on autosomal loci, the reciprocal deletion and duplication tended to have the frequency ratio of 2:1 (166), although the predominance of interchromosomal rearrangements in velocardiofacial syndrome (VCFS; MIM 192430) suggests a more equal ratio. Notably, the assays of Turner et al. (166) only measured the NAHR events across known meiotic recombination hotspots.
Lam & Jeffreys (75, 76) also performed pooled sperm assays on the alpha-globin locus in two subjects. Instead of measuring across hotspots in the LCR, they examined the entire globin locus and could thus also record NAHR events outside the hotspots and other non-NAHR rearrangements. In their study, one person showed the same deletion and duplication frequency, whereas the other one had more duplication than deletion. This apparent discrepancy between the two studies could be due to experimental design or it could reflect true differences among different NAHR loci, potentially influenced by the local genomic architecture.
Eichler and colleagues designed an aCGH platform to interrogate genomic regions with architectural features resulting in susceptibility to NAHR. Bioinformatic analyses of the human genome reference sequence were used to identify genomic intervals with substrate requirements (LCRs/SDs located on the same chromosome, directly oriented, and with high sequence identity) for NAHR (88). They then designed arrays to interrogate NAHR susceptible regions to investigate the genomic DNA from patients with mental retardation (MR) and/or multiple congenital anomalies (MCA) (143). In less than two years, they defined six new MR/MCA genomic disorders including microdeletions and/or microduplications of 1q21.1 (106), 15q13.3 (52, 144), 15q24 (145), 16p13.11 (50), 17q12 (105), and 17q21.31 (143). These and other studies clearly demonstrate the importance of understanding the mechanisms for human genomic rearrangements.
Some Simple Copy Number Variations Can Be Explained by Nonhomologous End-Joining
Nonhomologous end-joining (Figure 3) is utilized by human cells to repair DNA doublestrand breaks (DSBs) caused by ionizing radiation or reactive oxygen species and for physiological V(D)J recombination (83, 84, 136). Two characteristic features of NHEJ are: (a) Unlike NAHR, NHEJ does not require substrates with extended homology; (b) NHEJ can leave an ‘information scar’ in the form of loss or addition of several nucleotides at the join point (82).
Breakpoints of NHEJ-mediated rearrangements often fall within repetitive elements such as LTR, LINE, Alu, MIR, and MER2 DNA elements (120, 164). Moreover, sequence motifs like TTTAAA, known to be capable of causing DSB or curving DNA, are present in proximity to many of these junctions (120, 148, 164).
Also, many 17p translocations and other nonrecurrent deletions have one of their breakpoints in LCRs (155). These data suggested that although without any obligatory substrate requirement for LCRs, NHEJ may still be stimulated by certain genomic architectures (147, 155).
A DNA Replication–Based Mechanism, FoSTeS, Can Account for Complex Genomic Rearrangements and Copy Number Variations
Recently, the observation of the often complex details of genomic rearrangements, enabled by aCGH techniques, led Lee et al. (78) to propose the replication FoSTeS model as a mechanism for human genomic rearrangements (Figure 3).
According to this model, the DNA replication fork can stall, the lagging strand disengages from the original template and switches to another replication fork and restarts DNA synthesis on the new fork by priming it via the microhomology between the switched template site and the original fork (78). The new template strand is not necessarily adjacent to the original replication fork in primary sequence, but likely in three-dimensional physical proximity. Upon annealing, the transferred strand primes its own template-driven extension at the transferred fork. Depending on the direction of the fork progression and whether the lagging or leading strand in the new fork was used as a template and copied, the erroneously incorporated fragment from the new replication fork could be in direct or inverted orientation to its original position. Furthermore, depending on whether the new fork is located downstream or upstream of the original fork, the template switching results in either a deletion or a duplication. This procedure of disengaging, invading, and synthesizing may occur multiple times in series (i.e., FoSTeS × 2, FoSTeS × 3, etc.), likely reflecting the poor processivity of the DNA polymerase involved, and resulting in the observed complex rearrangements.
Array CGH data on genomic regions including the Pelizaeus-Merzbacher disease locus (78), the SMS/PTLS locus (130, 171, 186), the MECP2 locus (8, 22), the LIS1 locus (11), and a number of other loci have revealed the complex nature of many other nonrecurrent rearrangements, some of which were thought to be simple deletion or tandem duplication before the analysis by higher-resolution genome technologies. In addition, some complex chromosomal rearrangements unveiled by recent cytogenetic experiments can also be better explained by FoSTeS (171, 185).
In our studies on breakpoint sequences of FoSTeS-mediated complex rearrangements, e.g., complex LIS1 duplications (11), microhomologies shared between two Alu elements have also been identified at one or more breakpoints. Thus, in addition to Alu-Alu-mediated NAHR resulting in simple genomic rearrangement (2, 49, 139), microhomology shared between Alu elements can also apparently be used for template switching and priming DNA replication on a new fork. Since the prevalence of Alu elements has been observed at the ends of LCRs/SDs (6) and at breakpoints of subunits of complex LCRs/SDs (2), FoSTeS events involving Alu elements may also be responsible for LCR/SD formation.
Not only can FoSTeS generate large genomic duplications of several megabases (11, 78), it also causes genic duplication/triplication and even rearrangements of single exons (186), which implicate FoSTeS in gene duplication and exon shuffling; two predominant mechanisms driving gene and genome evolution (42, 121).
By reanalyzing the breakpoint sequence data of 23 deletion CNVs published by Perry et al. (125), we have found that at least 5 (22%) CNV breakpoints were highly complex and consistent with two or more FoSTeS events. Furthermore, many of the remaining deletion CNVs show microhomologies at breakpoints, which are also consistent with FoSTeS × 1 (125).
Based upon observations regarding genomic rearrangements in human (FoSTeS), bacteria, yeast, and other model organisms, Hastings et al. (51) proposed a generalized replicative, template-switch model that may underlie many structural variations in genomes and genes from all domains of life. This is termed the microhomology-mediated break-induced replication (MMBIR) model. MMBIR could not only lead to CNV by itself, but also create LCRs that provide the homology required for NAHR and predispose to more genomic rearrangements in future generations. MMBIR may cause somatic events associated with cancer and underlie the genomic rearrangements and CNV associated with the emergence of primate-specific traits providing variation for natural selection and evolution (51).
L1 Retrotransposition Contributes to Copy Number Variation
Long interspersed element-1 (L1) elements cover 16.89% of human genomic DNA and are the only currently active autonomous transposons in the human genome (45, 63). Of the 516,000 copies of L1 in our genome, only about 80–100 copies are full length (about 6 kb in size) and have two intact open reading frames (ORF): ORF1 coding for a RNA-binding protein and ORF2 encoding a protein with both endonuclease (EN) and reverse transcriptase (RT) activity (3).
L1 transposition occurs via an RNA intermediate that is probably transcribed by RNA polymerase II (3). The reverse transcription and integration are thought to occur in a coupled process called target primed reverse transcription (TPRT) (123) (Figure 3). The resultant insertion is flanked by duplicated target sites (TSD), characteristic of TPRT (123).
L1s are also responsible for the mobilization of Alu elements and SVA elements (SINER, VNTR) (3, 45, 122) as well as retrogenes. Korbel et al. attributed 30% of the SV indels that they detected to retrotransposition; of these 90% were due to L1 elements themselves and 8% to SVAs (68). Kidd et al. found 15% of the SV events that they detected to be due to retrotransposition (64). In the analysis of the mobile element-associated SVs in the Venter genome, Xing et al. found that ~10% of the indels of > 100 bp are associated with transposable DNA sequences, including L1, Alu, and SVA (181).
Almost all the sequenced SV breakpoints (N = 114) in Korbel et al. (68) bear signatures of either NAHR (surrounded by LCRs or other repetitive sequences), NHEJ/FoSTeS × 1 (microhomology at the junction), or retrotransposition (mostly L1 elements). Kidd et al. (64) also observed similar evidence at breakpoints that can be interpreted as having occured by NAHR, NHEJ, FoSTeS, and retrotransposition mechanisms. Thus, these mechanisms can explain the majority of the DNA rearrangements occurring in our genomes.
COPY NUMBER VARIATION: MUTATION RATES
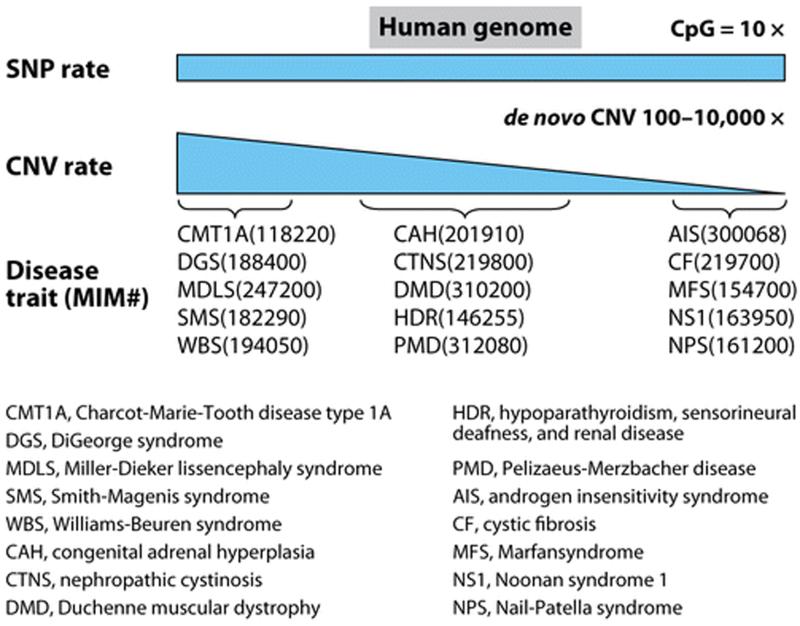
The mutation rate of point mutations is estimated to be 1.8–2.5 × 10−8 per base pair per generation (67, 115). Although precise estimates of CNV mutation rates are elusive, the locus-specific mutation rates of some de novo CNVs have been estimated by different methods, including studies of a single X-linked gene (DMD) (169), prevalence calculations, pooled sperm PCR assays, and aCGH analyses of trios. The estimates for CNV locus-specific mutation rates range from 1.7 × 10−6 to 1.0 × 10−4 per locus per generation (92, 169), i.e., 100 to 10,000 times higher than nucleotide substitution rates! Distinct from the relatively constant mutation rates of SNPs across the human genome [with the exception of a transition point mutation causing SNPs at CpG dinucleotides (19)], mutation rates of CNVs can vary widely at different loci, likely reflecting the differences in CNV formation mechanism and local or regional genome architecture inciting genomic instability (Figure 4).
Figure 4.
New mutation rates for SNP versus CNV. Constant SNP mutation rates and variable CNV mutation rates across the human genome. Examples of human disease traits (OMIM numbers are shown) with different contribution of CNV versus SNP. Note that for some diseases at specific loci, CNVs outweigh SNPs as a mutational cause for disease. CNV de novo locus-specific rates can vary throughout the genome, whereas SNP rates are essentially constant. The SNP rate for CpG dinucleotides is constant but about ten times higher than other bases because of methyl-mediated deamination of cytosine to uracil, causing transition mutations.
For autosomal dominant genomic disorders that do not result in embryonic lethality and are fully penetrant with fitness w, the per-locus per-generation rate (μ) of loss-of-function mutation can be estimated by the birth prevalence B, i.e., μ = (1/2)(1 – w)B (92). According to the assumed fitness of zero and the estimated birth prevalence, e.g., 1/4000 for DiGeorge syndrome (DGS)/VCFS at 22q11.2, 1/10000 for WBS at 7q11.23, and 1/25000 for SMS at 17p11.2 (142), the estimated mutation rates for these genomic rearrangements are 2 × 10−5 to 1.25 × 10−4 per locus (92). As determined by pooled sperm PCR assays directly in the male germline, the average rate for deletion CNV is 4.20 × 10−5 for the CMT1A locus, 9.55 × 10−6 for the WBS locus, 1.87 × 10−6 for the LCR17p locus, and 2.16 × 10−5 for the Y-linked AZFa locus (166). Notably, the mutation rate of CMT1A duplication based on the experimentally determined pooled sperm PCR assay (1.73 ± 0.49 × 10−5) (166) correlates very well with that determined theoretically from prevalence calculations (1.7 – 2.6 × 10−5) (92).
The overall rate of de novo CNV can also be explored via the genome-wide screening for de novo CNVs in trios. By reanalyzing the oligonucleotide genome-wide tiling-path aCGH data of Sebat et al. (137) from trios of autism cases and controls, the per-locus mutation rate (μ) was estimated by μ × 3 × 103 = 1/2 (1 × 10−2), i.e., μ = 1.7 × 10−6, based on the observations of 2 de novo CNVs (~1%) out of 196 controls and average CNV size of ~2 × 106 bp in the autism cohort (i.e., 2 × 106 such loci in the whole genome) (92). However, as the resolution limit of aCGH employed in Sebat et al. (137) prevents the detection of numerous small CNVs, the calculated CNV mutation rate of 1.7 × 10−6 from their data may thus be an underestimate. Note also that somatic mutation may contribute to some of these observations, and it is important to demonstrate germline transmission to distinguish germline from somatic variants.
With the aid of accumulating CNV data, more accurate mutation rates for CNV will be accessible. Nevertheless, existing data suggest the de novo rates for CNV may be orders of magnitude greater than for SNPs in some regions of the genome. However, given their dependency on genomic architecture, locus-specific CNV mutation rates are much more variable across the genome than are SNPs. The variable mutation rates of CNVs may, in part, cause the relative contribution of SNPs/CNVs to vary widely among Mendelian loci (Figure 4).
COPY NUMBER VARIATION: CONVEYED PHENOTYPES
Large chromosomal aberrations have long been known to be associated with human diseases. The best-known example is Down syndrome (MIM 190685) caused by trisomy of human chromosome 21 (79). Such large chromosome abnormalities are detectable by conventional microscopy. Copy number changes caused by submicroscopic genomic deletions were subsequently found to be involved in human diseases and other traits including thalassaemia due to alpha globin gene rearrangements (53) and red-green color blindness; the latter affects approximately 8% of Caucasian males. In 1986, the red-green pigment genes were mapped to Xq28 as a tandem array of one gene encoding red opsin and one or more genes encoding green opsin (118). The high degree of sequence similarity between these genes makes them vulnerable to NAHR and the consequent deletion or duplication polymorphisms, some of which can totally eliminate or disrupt red or green opsin genes and lead to red-green color blindness (117). Besides deletion resulting in defects in gene function, duplication involving a dosage-sensitive gene can also cause disease. In 1991, the first disease-associated submicroscopic duplications were identified in 17p12, and this duplication can lead to CMT1A (97, 99). Some examples of clinical phenotypes conveyed by CNVs involving single or multiple genes are shown in Table 2. Smaller CNVs affecting single exons also account for a proportion of human diseases (30, 73, 74).
Clinical findings associated with submicroscopic chromosomal imbalance (including deletions, duplications, insertions, translocations, and inversions) have been archived in DECIPHER (DatabasE of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources; http://www.sanger.ac.uk/PostGenomics/decipher/) (37). To date (May 2009), over 2,200 cases of >50 diseases have been included in DECIPHER.
CNV can be responsible for sporadic birth defects (87), other sporadic traits, Mendelian diseases, and complex traits (Table 2). In this review we expand on complex neurological diseases and susceptibility to complex diseases as sporadic and Mendelian disease have been reviewed elsewhere (88, 98, 154).
Complex Neurological Diseases
Parkinson disease
Parkinson disease (PD; MIM 168600) is a common neurodegenerative disorder with manifestations including resting tremor, muscular rigidity, bradykinesia, and postural instability that affects approximately 1% of the population over age 50 (128). At least 13 genomic loci have been reported to be involved in PD, including SNCA, the alpha-synuclein gene on 4q21 (128). Singleton et al. (150) identified a novel triplication involving SNCA linked to autosomal dominant PD in a large family, which implicates the dosage effect of SNCA in PD. In a further study of gene expression in the patients with the SNCA triplication, Singleton and colleagues, found an approximately twofold increase of SNCA protein in blood, a twofold increase of SNCA mRNA in brain tissue, and increased deposition of heavily aggregated SNCA protein in brain tissue (109).
Later studies also confirmed the role of SNCA copy number gain in PD. Chartier-Harlin et al. (17) discovered the SNCA triplication in one of nine families with autosomal dominant PD. SNCA triplication has also been reported in a family of Swedish American descent with autosomal dominant early-onset PD (32). In addition to the SNCA triplication, Fuchs et al. (41) also detected SNCA duplications in a Swedish family, which shares a common ancestor with the former reported family (32). Ibanez et al. (58) identified 2 with SNCA duplication out of 119 individuals from PD families. These observations strongly suggest a dosage effect of SNCA in selected cases of PD.
Alzheimer disease
Alzheimer disease (AD; MIM 104300) is a neurodegenerative disorder compromising cognition in the elderly, which is characterized by intracellular neurofibrillary tangles and extracellular amyloid plaques that accumulate in vulnerable brain regions (140). AD-associated point mutations have been identified in at least 15 genomic loci, including the APP gene encoding the amyloid precursor protein (43). Genetic variations in APP promoter sequences are also associated with AD. Theuns et al. (162) identified eight novel APP promoter variants in late-onset AD, three of which can cause a nearly twofold neuron-specific increase in APP transcriptional activity in vitro. This observation suggests that increased APP level can result in AD and that APP is a dosage-sensitive gene. Also of interest, Down syndrome due to trisomy 21 is associated with early-onset Alzheimer disease; the APP gene maps to chromosome 21 and so those with Down syndrome have three copies. Thus, copy number gain of APP is hypothesized to be one of the causes of AD. Duplication CNVs of the APP gene have been reported in families with autosomal dominant early-onset Alzheimer disease (ADEOAD) and cerebral amyloid angiopathy (CAA) (133). Rovelet-Lecrux et al. (133) identified five duplications of differing sizes and encompassing APP, which caused accumulation of beta-amyloid peptides and the consequent phenotype of ADEOAD with CAA. These APP duplications were present only in affected subjects and cosegregated with the disease in families and were not found in 100 healthy controls with normal cognition over the age of 60 years. APP duplications associated with dementia with CAA have also been identified in a Dutch population (151).
Mental retardation
Mental retardation (MR) is a nonprogressive cognitive impairment. Many de novo genomic rearrangements have been identified in patients with MR (21, 146, 153). Due to the relative small sample size in the early studies, these de novo CNVs were identified in single cases and each CNV was different (21, 146). Larger cohorts may help detect and confirm the roles of rare de novo CNVs in MR.
Recently, Froyen et al. (40) studied a large set of 300 well-characterized families with X-linked mental retardation (XLMR) and identified 6 overlapping duplications at Xp11.22 in 6 unrelated males with predominantly nonsyndromic XLMR. Their maximal common duplicated region is about 320 kb and involves four known genes (SMC1A, RIBC1, HSD17B10, and HUWE1), three candidates of which may convey the phenotype of MR. SMC1A encodes the subunit of cohesion complex, and point mutations in it can lead to Cornelia de Lange syndrome (MIM 300590) with facial dysmorphisms, MR, and growth deficits in childhood (112). A silent mutation of HSD17B10 has been reported to be a syndromic form of MR with choreoathetosis (80). In addition to the duplications involving HUWE1 (an E3 ubiquitin ligase gene), Froyen et al. (40) also identified three point mutations of the same gene in three XLMR families, a finding that highlights the role of HUWE1 in XLMR and supports the conclusion that it is a dosage-sensitive gene, at least partially responsible for the MR phenotype in the duplication case.
Loss-of-function mutations of the X-linked methyl-CpG-binding protein 2 gene (MECP2) at Xq28 are associated with developmental delay (DD), MR, and fatal infantile encephalopathy in males. Recent findings suggest increased MECP2 gene copy number can also convey a clinical phenotype, resulting in a DD/MR plus seizures phenotype in males (8, 15, 22, 168).
Recently, Bi et al. (11) identified seven unrelated cases with submicroscopic duplication in 17p13.3 involving the LIS1 and/or the 14-3-3ε genes and using a ‘reverse genomics’ approach characterized the clinical consequences of duplication. “Genomotype”/phenotype correlations showed that increased LIS1 dosage can cause microcephaly, mild brain structural abnormalities, moderate to severe developmental delay, and failure to thrive, whereas duplication of 14–3-3ε increases the risk for macrosomia, mild developmental delay, pervasive developmental disorder, and results in shared facial dysmorphologies.
Autism
Autism (MIM 209850) is a child psychiatric disorder that is characterized by a triad of limited or absent verbal communication, a lack of reciprocal social interaction or responsiveness, and restricted, stereotypical, and ritualized patterns of interests and behavior (4). To examine if CNVs can convey autism spectrum disorder (ASD), Sebat et al. (137) employed the ROMA technology in 165 families affected by autism and in 99 control families. They found significantly more spontaneous CNVs in ASD patients (14/195) than in unaffected controls (2/196) (P = 0.0005). Furthermore, the two CNVs in control subjects were duplications, whereas most of the CNVs (12/15) in ASD patients were deletions (137). These observations suggest that a high frequency of de novo CNVs in ASD patients, especially deletions, is a risk factor for autism. Some of the de novo CNVs identified by Sebat et al. (137) have been reported to be autism-associated or overlap with autism susceptibility (AUTS) loci, for example, the duplication of 15q11–13 (AUTS4; MIM 608636) and the deletion of 16p11.2 (AUTS14; MIM 611913).
The role of 16p11.2 deletion in autism was also confirmed in later studies. By reanalyzing the data of the genome-wide association study of 751 multiplex families from the Autism Genetic Resource Exchange (AGRE), Weiss et al. (175) observed five patients with a 593-kb de novo deletion on 16p11.2. Subsequent CGH revealed 5 more cases of 16p11.2 deletion in 512 children affected by DD, MR, or suspected ASD and 3 more deletions in 299 Icelandic patients affected by autism, whereas only 2 such deletions were identified in 18,834 unscreened Icelandic controls. Kumar et al. (70) discovered 2 de novo 16p11.2 deletions out of 180 autism probands but none in 372 controls. In the subsequent additional screening, they found a similar result (2 deletions in 532 probands and no deletion in 465 controls). These observations suggest a significant association of the 16p11.2 deletion with autism (P = 0.044). Weiss et al. (175) also documented the reciprocal duplications. Both the 16p11.2 deletion and its reciprocal duplication were shown to be risk factors for autism; CNV at 16p11.2 accounts for approximately 1% of autism cases (175). The association of reciprocal 16p11.2 deletion and duplication with autism was also confirmed by Marshall et al. (100) by a combined discovery of four 16p11.2 CNVs in 427 ASD patients versus none in 1652 controls (P = 0.002).
Using homozygosity mapping in pedigrees with shared ancestry, Morrow et al. (111) identified several large, inherited, homozygous deletions, including an 886-kb 3q24 deletion affecting DIA1, whose level of expression changes in response to neuronal activity.
Schizophrenia
Schizophrenia (MIM 181500) is a chronic, debilitating illness with both neurological and psychiatric features. It is common with a lifetime prevalence of approximately 1%. An increased number of CNVs were found to be associated with schizophrenia (173, 182).
In genome-wide studies of thousands of patients, two related studies, Stefansson et al. (156) and the International Schizophrenia Consortium (161), discovered new loci responsible for schizophrenia (Table 3). Stefansson et al. (156) first screened and identified 66 de novo CNVs in 9878 transmission sets, none of whom had been diagnosed with schizophrenia. Then they tested these de novo CNVs for disease association in a sample of 1433 patients with schizophrenia and related psychoses and 33,250 controls. Three deletions (located on 1q21.1, 15q11.2, and 15q13.3) were found to be nominally associated with schizophrenia and psychosis (156). The associations of these three deletions with schizophrenia were further confirmed by a follow-up investigation in a second sample of 3285 cases and 7951 controls. The International Schizophrenia Consortium (161) performed a genome-wide survey of rare CNVs in 3391 patients with schizophrenia and in 3181 ancestrally matched controls, and discovered two associated deletions on 1q21.1 and 15q13.3, which are identical to two of three deletions found in the cohort reported by Stefansson et al. (94, 156). Furthermore, both studies also replicated the previously reported 22q11.2 deletions with schizophrenia phenotype in DGS/VCFS (Table 3) (62). The International Schizophrenia Consortium (161) also documented that the total CNV load was greater in patients with schizophrenia versus controls.
Table 3.
Odds ratios for statistically significant associations of psychiatric illness with microdeletion of a given genomic intervala
| Data | Microdeletion | ||||
|---|---|---|---|---|---|
| Study | Cases | Controls | 22q11.2 (DGS/VCFS) | 15q13.3 | 1q21.1 |
| Stefansson et al. (156) | ~4000 | ~40,000 | ∞ | 14.83 | 11.54 |
| Int. SCZ Consortium (161) | 3391 | 3181 | 21.6 | 17.9 | 6.6 |
From Reference 94.
Vrijenhoek et al. (172) identified 90 CNVs in 54 patients with schizophrenia; 13 of these are rare and not yet reported in human populations. Some of these rare CNVs were found to disrupt schizophrenia-associated genes, such as MYT1L, CTNND2, and ASTN2 (172). Therefore, rare CNVs affecting functional genes can be an important causative variation resulting in complex diseases such as schizophrenia. However, given the low frequency of these rare CNVs, large sample sizes may be required to confirm the association with disease.
Susceptibility to Other Complex Traits
It has long been known that the deletion of the alpha-globin gene can lead to alpha-thalassaemia (53) and protect against malaria (38). Recently, many more CNVs have been reported to affect disease susceptibility.
HIV susceptibility
Chemokines are secreted proteins involved in immunoregulatory and inflammatory processes (116). The CCL3L1 gene on 17q12 encodes the potential ligand for CC chemokine receptor 5, the major coreceptor for HIV (107). Therefore, CCL3L1 can be a dominant HIV-suppressive chemokine (107). CNVs of CCL3L1 (from 0 to 10 copies) have been reported in the Caucasian population (165). Gonzalez et al. (44) examined the effects of the CCL3L1 CNVs on the susceptibility to HIV and showed that low CCL3L1 copy number (below population average) is associated with markedly enhanced HIV/AIDS susceptibility. Interestingly, a strong association was detected between higher infant CCL3L1 copies and reduced susceptibility to HIV in the absence of maternal nevirapine (69).
Crohn disease and psoriasis
Crohn disease (CD), an inflammatory bowel disease, is a chronic disorder that causes inflammation of the digestive tract (MIM 266600). It has been shown that deficient expression of defensins, endogenous antimicrobial peptides protecting intestinal mucosa against bacterial invasion, can lead to chronic CD (34). Therefore, it was hypothesized that low copy number of the beta-defensin gene cluster may also be associated with chronic CD (33). Fellermann et al. (33) showed a median of three HBD-2 (human beta-defensin 2) copies per genome in colonic CD patients, which was significantly lower than the median of four HBD-2 copies in healthy controls (P = 0.002). Individuals with three or lower copies of HBD-2 have a significantly higher risk of developing colonic CD than do individuals with four or more copies (odds ratio 3.06).
Different from the decreased risk of colonic CD in individuals with high beta-defensin gene copies, copy number gain of beta-defensin genes was shown to be associated with psoriasis (MIM 177900), a chronic inflammatory dermatosis that affects approximately 2% of the population (55).
SNPs around IRGM (immunity-related GTPase family, M) have been associated with CD (124, 176). Recently, McCarroll et al. (103) showed that a previously known 20-kb deletion polymorphism upstream of IRGM is in perfect linkage disequilibrium with a CD-associated IRGM SNP. The deletion haplotype of IRGM was shown to have a distinct expression pattern of IRGM compared to the reference haplotype. Given that the IRGM expression can affect cellular autophagy of internalized bacteria, it was suggested that the common deletion polymorphism of IRGM may cause CD through the altered level of IRGM expression, affecting the efficacy of autophagy (103).
Pancreatitis
In the 1990s, several genes were determined to be associated with pancreatitis (MIM 167800), including the cationic trypsinogen gene PRSS1 (178). Considering that the pancreatitis-associated missense mutations of PRSS1, for example, the R122H mutation, increase trypsin activity in vitro (134), it was hypothesized that PRSS1 is a dosage-sensitive gene. When investigating a cohort of 34 French families with hereditary pancreatitis, Le Marechal et al. (77) did not identify any known point mutations in the pancreatitis-associated genes, but a 605-kb triplication encompassing PRSS1 was discovered in five families, which represents an identical-by-descent mutation.
Systemic lupus erythematosus and glomerulonephritis
Systemic lupus erythematosus (SLE; MIM 152700) is a chronic, remitting, relapsing, inflammatory, and often febrile multisystemic disorder affecting skin, joints, kidneys, and serosal membranes, due to failure in regulation of the immune system.
In an analysis of Fc receptor polymorphisms in northern European nuclear families with SLE, Aitman and colleagues found unexpected Mendelian errors at the FCGR3B gene in 14% of these families, which was hypothesized to be caused by CNV (1). Therefore, Aitman et al. (1) examined the potential FCGR3B CNV in 30 individuals from 8 nuclear families, in which FCGR3B polymorphisms showed Mendelian errors, and identified significant variation in FCGR3B copy number. Though no association between FCGR3B CNV and SLE was detected, a weak association was identified with lupus nephritis, an SLE subgroup with glomerulonephritis. This association was further strengthened in a later study by these authors with a larger sample size (P value reduced from 1 × 10−3 to 1.4 × 10−8) (31). Furthermore, an increased risk for development of SLE in individuals with fewer than two copies of FCGR3B was reported in the U.K. cohort (31). Willcocks et al. (179) confirmed the association of SLE with low FCGR3B CNV in Caucasians.
Complement component 4 (C4, including C4A and C4B) gene mutations have long been known to be associated with SLE (36). Yang et al. (184) examined the C4 CNVs in 1241 Americans of European descent. The copy number of C4 varied from 2 to 6 (C4A, 0 to 5; C4B, 0 to 4), and the risk of SLE increased in the subjects with low C4 copies but decreased in those with high C4 copies (184).
Molecular Mechanisms by which Copy Number Variations Convey Phenotype
CNVs caused by genomic rearrangements can convey phenotypes by the following molecular mechanisms: (a) gene dosage, (b) gene interruption, (c) gene fusion, (d) position effects, (e) unmasking of recessive alleles or functional polymorphism, and (f) potential transvection effects (98) (Table 4).
Table 4.
Specific example of copy number variation (CNV) disease mechanisms
| Mechanism | Affected gene | Disease | Reference(s)a |
|---|---|---|---|
| Gene dosage | PMP22 | CMT1A/HNPP | S11, S40, S41, S60 |
| Gene interruption | F8 | Hemophilia A | S2 |
| Gene fusion |
CYP11B1,
CYP11B2 |
Hypertension and GRAb |
S39 |
| Position effects | SOX9 | Campomelic dysplasia |
S7, S66 |
| Unmasking of recessive alleles or functional polymorphism |
FXII | Sotos syndrome | S35 |
| Potential transvection effect | Rai1 | SMS | S73 |
For the list of supplemental references, follow the Supplemental Material link from the Annual Reviews home page at http://www.annualreviews.org.
GRA, glucocorticoid-remediable aldosteronism.
CNVs involving dosage-sensitive genes, such as PMP22, can alter gene expression levels and cause consequent clinical phenotypes. PMP22 (encoding peripheral myelin protein) is located within the 1.4-Mb CMT1A region at 17p12, whose duplication can lead to CMT1A by PMP22 overexpression (96, 97, 99), whereas deletion can result in HNPP by PMP22 under-expression (i.e., haploinsufficiency) (16, 96).
When the breakpoint of a deletion, insertion, or tandem duplication is located within a functional gene, it may interrupt the gene and cause a loss of function by inactivating a gene as exemplified by red-green opsin genes and color blindness (117). Gene fusion caused by genomic rearrangements between different genes or their regulatory sequences can generate a gain-of-function mutation. This mechanism is prominent among cancers associated with specific somatic chromosomal translocations. Disease-associated gene fusion has also been found in hypertension (85). Genes encoding aldosterone synthase and steroid 11 beta-hydroxylase on 8q are candidate genes for glucocorticoid-remediable aldosteronism (GRA, an autosomal dominant disorder that is characterized by hypertension with variable hyperaldosteronism). These two genes have 95% identity, and NAHR-caused gene fusion between them segregates with GRA in a large kindred (85).
By removing or altering a regulatory sequence, CNV can have an effect on expression or regulation of a nearby gene out of the CNV region, i.e., position effect (66). For example, mutations in SOX9 lead to campomelic dysplasia, but Velagaleti et al. (170) reported that two balanced translocations, with breakpoints mapping to approximately 900 kb upstream and 1.3 Mb downstream of SOX9 can also cause disease. Many other CNVs have been identified to alter gene expression and cause human diseases (66).
Deletion removing one allele may unmask another recessive allele or functional polymorphism. For instance, the activity of the plasma coagulation factor 12 (FXII) in patients with the common Sotos syndrome deletion is predominantly determined by the functional polymorphism of the remaining hemizygous FXII allele (71).
Transvection, the influence of gene expression by the pairing of alleles on homologous chromosomes, is one mechanism for trans regulation (25). Its effect is mediated via deletion of regulatory elements required for communication between alleles. When studying the mouse models of SMS, Yan et al. (183) found that the penetrance of craniofacial anomalies (a major clinical manifestation of SMS) was modified by the 590-kb genomic sequence surrounding Rai1, in which potential transvection or other trans-regulatory factors may exist.
COPY NUMBER VARIATION: EVOLUTION
CNVs can lead to diseases or other human traits by involving dosage-sensitive genes, disrupting functional genes, or other molecular mechanisms. Therefore, CNVs can also be potentially exposed to selection pressure during evolution, which has been confirmed by the observations in humans and other primates, mice, and fruit flies.
Purifying Selection
Substantial evidence that the location of CNVs is biased away from functional sequences has been found in human genomes, which suggests purifying selection on CNVs within humans. By determining the proportion of SNPs within deletion CNVs in coding sequence versus introns, Conrad et al. (18) found strong underrepresentation of genic SNPs in deletion regions compared with the HapMap average. This finding has been confirmed by the observations of Redon et al. (132) that CNVs are preferentially located outside of genes and ultraconserved elements in the human genome and that a significantly lower proportion of deletions than duplications overlaps with disease-related genes and RefSeq genes.
Purifying selection on CNVs, especially deletions, was also observed in flies. Emerson et al. (29) used genome-tiling arrays to study the CNVs in D. melanogaster and detected 2658 independent CNVs, where duplications outnumbered deletions (dup/del = 2.5). This indicated purifying selection on deletion CNVs (29). Dopman & Hartl (23) also detected a similar strong purifying selection on the CNVs in the Drosophila genome, especially those located in functionally constrained regions.
Gene Duplication and Positive Selection
Gene duplication has long been thought to be a central mechanism driving long-term evolutionary changes (56, 121). Selection has also been shown to shape the architecture of segmental duplications during human genome evolution (61). Studying CNVs, especially gene amplification favored by positive selection, during evolution may help us discover new functional genes and reveal the genomic alteration and environmental impact driving human evolution.
Sikela and colleagues (24) used aCGH of 41,126 human cDNA from 24,473 unique human genes to study gene CNVs spanning 60 million years of human and primate evolution. Gene duplications and losses were surveyed on a genome-wide scale across 10 primate species including human. It was found that 6,696 (27.4%) of the examined human genes represent CNVs in one or more of the 10 primate species (24). Remarkably, gene gains typically outnumbered losses (gains/losses = 2.34), which suggests positive selection in primate genome evolution. Furthermore, some CNVs discovered are lineage specific (LS) (24). Studying these human LS CNVs may reveal the evolutionary process driving the emergence of human-specific traits such as cognition.
In their earlier study of the human LS amplification (129), Sikela and colleagues found that the strongest signal comes from the multiple-copy protein domain, DUF1220. The copy number of DUF1220 was shown to be highly expanded in humans, reduced in African great apes, further reduced in orangutan and Old World monkeys, only single-copy in nonprimate mammals, and absent in nonmammalian species (129). Examination of expression in brain showed that neuron-specific DUF1220 signals were present in the cortical layers of the hippocampus and also abundant in neurons within the neocortex (129). Both evolutionary and functional evidence suggests DUF1220 and its expansion in the human lineage is critical to higher cognitive functions. Notably, the majority of DUF1220 sequences are located at 1q21.1 (129), lying within regions of CNV that are associated with MR (143), schizophrenia (156, 161, 173), and microcephaly/macrocephaly (13). This suggests a link between DUF1220 and human brain/cognition function and behavior.
Other examples of human LS gene copy-number expansions are AQP7. This is important for increasing glycerol transport for mobilization of energy stores and possibly water transport involved in exercise-induced sweating (24) and the salivary amylase gene, AMY1, which is correlated positively with salivary amylase protein levels and amount of starch in the diet among humans (126).
These examples of human-specific or primate-specific gene amplification illustrate that CNVs encompassing functional genes can be evolutionally favored because of their adaptive benefits. Similar evidence for positive selection on gene duplication has also been found in other species. In the study of Emerson et al. on flies, 56% of the CNVs were found to affect genes and most notably high-frequency duplication CNVs were found to involve toxin-response genes (for example, Cyp6g1 contributing to resistance to DTT). This suggested potential positive selection on these CNVs (29). In inbred mouse strains, large and complex interstrain CNVs (mainly duplications) were shown to be restricted to gene families functional in spermatogenesis, pregnancy, and immune response (149). However, it was recently shown that the genic biases of CNVs could alternatively be explained by reduced efficiency of purifying selection in eliminating deleterious changes in humans (119). In addition to gene duplication, CNV can also lead to exon shuffling (42, 186), another hypothesis proposed to be responsible for the origin of new genes.
CONCLUSION
CNV has been recognized as a predominant source of genetic variation among human individuals. CNV encompasses more human genomic content and has a higher per-locus mutation rate than does SNP. Recombination-based mechanisms (e.g., NAHR and NHEJ), retrotransposition, and a new replication-based FoSTeS and/or MMBIR mechanism have been shown to play roles in CNV formation. CNV can convey sporadic diseases, Mendelian and complex traits by dosage effects [triplication, e.g., PLP1 (180), MECP2 (22), LIS1 (11), sometimes conveying more severe phenotypic consequences than duplication], gene disruption, position effect, and other molecular mechanisms. CNV is also thought to drive human genome evolution via gene duplication and exon shuffling. However, due to the limits of interrogating resolution and genome coverage of the present technologies employed in CNV studies, much more undiscovered CNV may exist in the human genomes, and further comprehensive investigation is expected to advance our knowledge of the distribution, formation, conveyed phenotype or genetic susceptibility, selection, and evolution of CNV.
Supplementary Material
SUMMARY POINTS.
Structural variation, including CNV, is responsible for a large fraction of human genetic variation.
CNVs can represent benign polymorphic variations or convey clinical phenotypes by mechanisms such as altered gene dosage and gene disruption.
Human genome rearrangements can occur by several mechanisms that include both recombination (NAHR and NHEJ) and replication (FoSTeS/MMBIR)-based mechanisms. The latter can result in complex genomic rearrangements.
Locus-specific de novo mutation rates for CNV can be 100 to 10,000 times more frequent than for SNP.
The relative contribution of CNV to locus-specific mutation rate may vary throughout the human genome and can reflect local genome architecture resulting in regional susceptibility to genome instability.
CNV is important to human genome and gene evolution.
Gene duplication and triplication, and potentially exon shuffling, can occur by the FoS-TeS and/or MMBIR mechanism.
FUTURE ISSUES.
How should we define CNV? CNVs have been defined as “a segment of DNA that is 1 kb or larger and is present at a variable copy number in comparison with a reference genome” (35). However, the cutoff of 1 kb is completely arbitrary. In fact, one might argue that 2 bp or more is a CNV versus a SNP using a chemical definition that SNP changes only the base in the DNA, whereas the sugar-phosphate backbone needs to be disrupted/altered to make a CNV. Nevertheless, based on a functional definition, it may be better to choose an average exon size (~100 bp) as a parameter for defining CNV. Recent observations in the Watson and Venter genomes clearly indicate that the CNV size distributions show a marked enrichment in the range of 300 to 350 bp owing to the known retrotransposition-based Alu polymorphisms (81, 177) (Figure 1). As analyses of higher resolution are applied, many more CNVs of smaller size ranges are likely to be discovered (Figure 1).
What will be the standard for determining if a copy number variant causes or is associated with susceptibility to a given phenotype? It is challenging to ascribe a phenotype to a CNV [genomotype/phenotype correlations (11)] because often there is no genetic code to help establish causation as there can be for nucleotide changes that result in nonsense or frameshift alleles. Penetrance for a CNV may not be complete and duplication CNVs may confer milder phenotypes than deletion CNVs at a given locus. Finally, it is extremely difficult to determine that a CNV does not influence a phenotype, challenging the notion of CNVs as benign polymorphic variants. Bringing together the common disease genetics community, which is undertaking large-scale association studies, and the clinical genetics community, which seeks to generate meaningful results for individual patients with relatively rare phenotypes, into a unified view of the allelic architecture of genetic diseases must be a clear objective for the coming years.
Will forward and reverse genomics identify the molecular bases of many traits? For almost a century forward genetics has been used to map Mendelian traits to specific loci in many organisms. With the advent of recombinant DNA, reverse genetics has enabled the creation of specific gene mutations and the elucidation of the phenotypic consequences of single gene alterations. Currently, the forward genomics clinical implementation of genome-wide arrays to identify CNVs responsible for disease traits is uncovering many regions of the human genome wherein genomic changes result in disease or susceptibility to disease. Can reverse genomics now be used to systematically analyze phenotype(s) conveyed by either a deletion or duplication of a specific region of the human genome? To what extent will reverse genomics help elucidate gene function for the remaining ~ 90% of annotated genes not yet assigned a function and also address the question: What is the genomic code (95)?
Will CNV provide novel routes to therapy? If a CNV can result in a disease phenotype by virtue of gene dosage, will normalizing gene dosage through epigenetic manipulation correct the disease (93)? Studies of an animal model for the CMT1A duplication demonstrate that the neuropathy phenotype due to PMP22 overexpression can be mitigated by treatment with a progesterone antagonist that reduces PMP22 expression (108, 141). Perhaps RNAi may be effective in reducing the expression of the dosage sensitive gene(s) in duplication disorders.
What will be the impact of CNV on evolution? Due to the low resolution of methods utilized previously (e.g., BAC aCGH) in CNV screening, the majority of the identified CNVs have not yet been finely resolved to the nucleotide level. This precludes the ability to perform CNV genotyping. For over 90% of reported CNVs, we do not know their true population frequency because they have not been genotyped. To study the roles of CNV in genome evolution, we will need large CNV genotype datasets from different populations. Also, precise de novo CNV mutation rates throughout the genome are required to better understand the contribution of CNV versus SNP to genome evolution, particularly with respect to gene duplication/triplication and exon shuffling.
We need a better reference human genome. For designing genome-wide arrays and comparisons of personal genome sequences, one requires some reference from which to choose oligonucleotides to be placed on arrays and also to use for computational comparative analysis when sequencing. The current draft/finished genome must have its gaps filled and robust sequences for both heterochromatic transition regions and heterochromatin determined. Furthermore, CNV information needs to be integrated with the reference sequence such that nucleotide positions of breakpoints, rather than just genomic regions, are detailed in the reference genome. Comparison with the current reference genome can also lead to erroneous conclusions. For example, we have experienced in the studies on genomic duplications at the SMS locus that a de novo tandem duplication spanning across one end of an inversion allele can be misinterpreted to be a de novo complex duplication because only one inversion haplotype is in the current reference genome (186). Also, as mentioned above, personal genome sequencing may identify genomic segments not present on the current reference sequence. Thus, because the current reference genome sequence contains the deleted allele at a number of common CNVs, these will be missed by microarray-based surveys given that the array design utilizes the reference genome sequence.
ACKNOWLEDGMENTS
We thank Drs. Nicola Bruneti-Pieri, Claudia Carvalho, Katrina Gwinn, and Pawel Stankiewicz for their critical reviews. Work in the Lupski laboratory has been sponsored by the National Institutes of Health, the March of Dimes, the Muscular Dystrophy Association, the Foundation Fighting Blindness, and the Charcot-Marie-Tooth Association. Work in the Hurles laboratory is supported by the Wellcome Trust. Wenli Gu is a Feodor-Lynen Research Fellow generously supported by the Alexander-von-Humboldt Stiftung.
Glossary
- CNV
copy number variation
- SV
structural variation
- NAHR
nonallelic homologous recombination
- NHEJ
nonhomologous end-joining
- FoSTeS
fork stalling and template switching
- ROMA
representational oligonucleotide microarray analysis
- BAC
bacterial artificial chromosome
- DGV
Database of Genomic Variants
- aCGH
array comparative genomic hybridization
- PEM
paired-end mapping
- LCR
low-copy repeat
- PRIS
potential recombinogenic inverted sequences
- SD
segmental duplication
- MCA
multiple congenital anomalies
- MMBIR
microhomology-mediated break-induced replication
- ASD
autism spectrum disorder
Footnotes
DISCLOSURE STATEMENT
J.R.L. is a consultant for Athena Diagnostics, 23andMe, and Ion Torrent Systems Inc., and holds multiple U.S. and European patents for DNA diagnostics. Furthermore, the Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from molecular diagnostic testing (MGL, http://www.bcm.edu/geneticlabs/).
LITERATURE CITED
- 1.Aitman TJ, Dong R, Vyse TJ, Norsworthy PJ, Johnson MD, et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–55. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- 2.Babcock M, Pavlicek A, Spiteri E, Kashork CD, Ioshikhes I, et al. Shuffling of genes within low-copy repeats on 22q11 (LCR22) by Alu-mediated recombination events during evolution. Genome Res. 2003;13:2519–32. doi: 10.1101/gr.1549503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babushok DV, Kazazian HH., Jr. Progress in understanding the biology of the human mutagen LINE-1. Hum. Mutat. 2007;28:527–39. doi: 10.1002/humu.20486. [DOI] [PubMed] [Google Scholar]
- 4.Bailey A, Phillips W, Rutter M. Autism: towards an integration of clinical, genetic, neuropsychological, and neurobiological perspectives. J. Child Psychol. Psychiatry. 1996;37:89–126. doi: 10.1111/j.1469-7610.1996.tb01381.x. [DOI] [PubMed] [Google Scholar]
- 5.Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–7. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 6.Bailey JA, Liu G, Eichler EE. An Alu transposition model for the origin and expansion of human segmental duplications. Am. J. Hum. Genet. 2003;73:823–34. doi: 10.1086/378594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbouti A, Stankiewicz P, Nusbaum C, Cuomo C, Cook A, et al. The breakpoint region of the most common isochromosome, i(17q), in human neoplasia is characterized by a complex genomic architecture with large, palindromic, low-copy repeats. Am. J. Hum. Genet. 2004;74:1–10. doi: 10.1086/380648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauters M, Van Esch H, Friez MJ, Boespflug-Tanguy O, Zenker M, et al. Nonrecurrent MECP2 duplications mediated by genomic architecture-driven DNA breaks and break-induced replication repair. Genome Res. 2008;18:847–58. doi: 10.1101/gr.075903.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bayes M, Magano LF, Rivera N, Flores R, Perez Jurado LA. Mutational mechanisms of Williams-Beuren syndrome deletions. Am. J. Hum. Genet. 2003;73:131–51. doi: 10.1086/376565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bi W, Sapir T, Shchelochkov OA, Zhang F, Withers MA, et al. Increased LIS1 expression affects human and mouse brain development. Nat. Genet. 2009;41:168–77. doi: 10.1038/ng.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bovee D, Zhou Y, Haugen E, Wu Z, Hayden HS, et al. Closing gaps in the human genome with fosmid resources generated from multiple individuals. Nat. Genet. 2008;40:96–101. doi: 10.1038/ng.2007.34. [DOI] [PubMed] [Google Scholar]
- 13.Brunetti-Pierri N, Berg JS, Scaglia F, Belmont J, Bacino CA, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat. Genet. 2008;40:1466–71. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carvalho CM, Lupski JR. Copy number variation at the breakpoint region of isochromosome 17q. Genome Res. 2008;18:1724–32. doi: 10.1101/gr.080697.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carvalho CM, Zhang F, Liu P, Patel P, Sahoo T, et al. Some complex rearrangements in patients with duplication of MECP2 may occur by fork stalling and template switching. Hum. Mol. Genet. 2008;18:2188–203. doi: 10.1093/hmg/ddp151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chance PF, Alderson MK, Leppig KA, Lensch MW, Matsunami N, et al. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell. 1993;72:143–51. doi: 10.1016/0092-8674(93)90058-x. [DOI] [PubMed] [Google Scholar]
- 17.Chartier-Harlin MC, Kachergus J, Roumier C, Mouroux V, Douay X, et al. Alpha-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet. 2004;364:1167–69. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 18.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat. Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 19.Cooper DN, Youssoufian H. The CpG dinucleotide and human genetic disease. Hum. Genet. 1988;78:151–55. doi: 10.1007/BF00278187. [DOI] [PubMed] [Google Scholar]
- 20.Cusco I, Corominas R, Bayes M, Flores R, Rivera-Brugues N, et al. Copy number variation at the 7q11.23 segmental duplications is a susceptibility factor for the Williams-Beuren syndrome deletion. Genome Res. 2008;18:683–94. doi: 10.1101/gr.073197.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Vries BB, Pfundt R, Leisink M, Koolen DA, Vissers LE, et al. Diagnostic genome profiling in mental retardation. Am. J. Hum. Genet. 2005;77:606–16. doi: 10.1086/491719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.del Gaudio D, Fang P, Scaglia F, Ward PA, Craigen WJ, et al. Increased MECP2 gene copy number as the result of genomic duplication in neurodevelopmentally delayed males. Genet. Med. 2006;8:784–92. doi: 10.1097/01.gim.0000250502.28516.3c. [DOI] [PubMed] [Google Scholar]
- 23.Dopman EB, Hartl DL. A portrait of copy-number polymorphism in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2007;104:19920–25. doi: 10.1073/pnas.0709888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dumas L, Kim YH, Karimpour-Fard A, Cox M, Hopkins J, et al. Gene copy number variation spanning 60 million years of human and primate evolution. Genome Res. 2007;17:1266–77. doi: 10.1101/gr.6557307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duncan IW. Transvection effects in Drosophila. Annu. Rev. Genet. 2002;36:521–56. doi: 10.1146/annurev.genet.36.060402.100441. [DOI] [PubMed] [Google Scholar]
- 26.Eichler EE. Widening the spectrum of human genetic variation. Nat. Genet. 2006;38:9–11. doi: 10.1038/ng0106-9. [DOI] [PubMed] [Google Scholar]
- 27.Eichler EE, Clark RA, She X. An assessment of the sequence gaps: unfinished business in a finished human genome. Nat. Rev. Genet. 2004;5:345–54. doi: 10.1038/nrg1322. [DOI] [PubMed] [Google Scholar]
- 28.Eichler EE, Nickerson DA, Altshuler D, Bowcock AM, Brooks LD, et al. Completing the map of human genetic variation. Nature. 2007;447:161–65. doi: 10.1038/447161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emerson JJ, Cardoso-Moreira M, Borevitz JO, Long M. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science. 2008;320:1629–31. doi: 10.1126/science.1158078. [DOI] [PubMed] [Google Scholar]
- 30.Erlandson A, Samuelsson L, Hagberg B, Kyllerman M, Vujic M, Wahlstrom J. Multiplex ligation-dependent probe amplification (MLPA) detects large deletions in the MECP2 gene of Swedish Rett syndrome patients. Genet. Test. 2003;7:329–32. doi: 10.1089/109065703322783707. [DOI] [PubMed] [Google Scholar]
- 31.Fanciulli M, Norsworthy PJ, Petretto E, Dong R, Harper L, et al. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat. Genet. 2007;39:721–23. doi: 10.1038/ng2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farrer M, Kachergus J, Forno L, Lincoln S, Wang DS, et al. Comparison of kindreds with parkinsonism and alpha-synuclein genomic multiplications. Ann. Neurol. 2004;55:174–79. doi: 10.1002/ana.10846. [DOI] [PubMed] [Google Scholar]
- 33.Fellermann K, Stange DE, Schaeffeler E, Schmalzl H, Wehkamp J, et al. A chromosome 8 gene-cluster polymorphism with low human beta-defensin 2 gene copy number predisposes to Crohn disease of the colon. Am. J. Hum. Genet. 2006;79:439–48. doi: 10.1086/505915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fellermann K, Wehkamp J, Herrlinger KR, Stange EF. Crohn’s disease: a defensin deficiency syndrome? Eur. J. Gastroenterol. Hepatol. 2003;15:627–34. doi: 10.1097/00042737-200306000-00008. [DOI] [PubMed] [Google Scholar]
- 35.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat. Rev. Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 36.Fielder AH, Walport MJ, Batchelor JR, Rynes RI, Black CM, et al. Family study of the major histocompatibility complex in patients with systemic lupus erythematosus: importance of null alleles of C4A and C4B in determining disease susceptibility. Br. Med. J. 1983;286:425–28. doi: 10.1136/bmj.286.6363.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, et al. DECIPHER: DatabasE of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–33. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flint J, Hill AV, Bowden DK, Oppenheimer SJ, Sill PR, et al. High frequencies of alpha-thalassaemia are the result of natural selection by malaria. Nature. 1986;321:744–50. doi: 10.1038/321744a0. [DOI] [PubMed] [Google Scholar]
- 39.Flores M, Morales L, Gonzaga-Jauregui C, Dominguez-Vidana R, Zepeda C, et al. Recurrent DNA inversion rearrangements in the human genome. Proc. Natl. Acad. Sci. USA. 2007;104:6099–106. doi: 10.1073/pnas.0701631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Froyen G, Corbett M, Vandewalle J, Jarvela I, Lawrence O, et al. Submicroscopic duplications of the hydroxysteroid dehydrogenase HSD17B10 and the E3 ubiquitin ligase HUWE1 are associated with mental retardation. Am. J. Hum. Genet. 2008;82:432–43. doi: 10.1016/j.ajhg.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fuchs J, Nilsson C, Kachergus J, Munz M, Larsson EM, et al. Phenotypic variation in a large Swedish pedigree due to SNCA duplication and triplication. Neurology. 2007;68:916–22. doi: 10.1212/01.wnl.0000254458.17630.c5. [DOI] [PubMed] [Google Scholar]
- 42.Gilbert W. Why genes in pieces? Nature. 1978;271:501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- 43.Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349:704–6. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 44.Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–40. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 45.Goodier JL, Kazazian HH., Jr. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 46.Greenawalt DM, Cui X, Wu Y, Lin Y, Wang HY, et al. Strong correlation between meiotic crossovers and haplotype structure in a 2.5-Mb region on the long arm of chromosome 21. Genome Res. 2006;16:208–14. doi: 10.1101/gr.4641706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gu W, Zhang F, Lupski JR. Mechanisms for human genomic rearrangements. PathoGenetics. 2008;1:4. doi: 10.1186/1755-8417-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han K, Lee J, Meyer TJ, Remedios P, Goodwin L, Batzer MA. L1 recombination-associated deletions generate human genomic variation. Proc. Natl. Acad. Sci. USA. 2008;105:19366–71. doi: 10.1073/pnas.0807866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han K, Lee J, Meyer TJ, Wang J, Sen SK, et al. Alu recombination-mediated structural deletions in the chimpanzee genome. PLoS Genet. 2007;3:1939–49. doi: 10.1371/journal.pgen.0030184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hannes FD, Sharp AJ, Mefford HC, de Ravel T, Ruivenkamp CA, et al. Recurrent reciprocal deletions and duplications of 16p13.11: The deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. J. Med. Genet. 2009;46:223–32. doi: 10.1136/jmg.2007.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Helbig I, Mefford HC, Sharp AJ, Guipponi M, Fichera M, et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat. Genet. 2009;41:160–62. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Higgs DR, Pressley L, Old JM, Hunt DM, Clegg JB, et al. Negro alpha-thalassaemia is caused by deletion of a single alpha-globin gene. Lancet. 1979;2:272–76. doi: 10.1016/s0140-6736(79)90290-3. [DOI] [PubMed] [Google Scholar]
- 54.Hinds DA, Kloek AP, Jen M, Chen X, Frazer KA. Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat. Genet. 2006;38:82–85. doi: 10.1038/ng1695. [DOI] [PubMed] [Google Scholar]
- 55.Hollox EJ, Huffmeier U, Zeeuwen PL, Palla R, Lascorz J, et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nat. Genet. 2008;40:23–25. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hurles M. Gene duplication: the genomic trade in spare parts. PLoS Biol. 2004;2:e206. doi: 10.1371/journal.pbio.0020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, et al. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–51. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 58.Ibanez P, Bonnet AM, Debarges B, Lohmann E, Tison F, et al. Causal relation between alpha-synuclein gene duplication and familial Parkinson’s disease. Lancet. 2004;364:1169–71. doi: 10.1016/S0140-6736(04)17104-3. [DOI] [PubMed] [Google Scholar]
- 59.Int. Human Genome Sequencing Consort. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–45. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 60.Jeffreys AJ, Neumann R, Panayi M, Myers S, Donnelly P. Human recombination hot spots hidden in regions of strong marker association. Nat. Genet. 2005;37:601–6. doi: 10.1038/ng1565. [DOI] [PubMed] [Google Scholar]
- 61.Jiang Z, Tang H, Ventura M, Cardone MF, Marques-Bonet T, et al. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat. Genet. 2007;39:1361–68. doi: 10.1038/ng.2007.9. [DOI] [PubMed] [Google Scholar]
- 62.Karayiorgou M, Morris MA, Morrow B, Shprintzen RJ, Goldberg R, et al. Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc. Natl. Acad. Sci. USA. 1995;92:7612–16. doi: 10.1073/pnas.92.17.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kazazian HH, Jr, Moran JV. The impact of L1 retrotransposons on the human genome. Nat. Genet. 1998;19:19–24. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- 64.Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim PM, Lam HY, Urban AE, Korbel JO, Affourtit J, et al. Analysis of copy number variants and segmental duplications in the human genome: evidence for a change in the process of formation in recent evolutionary history. Genome Res. 2008;18:1865–74. doi: 10.1101/gr.081422.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kondrashov AS. Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Hum. Mutat. 2003;21:12–27. doi: 10.1002/humu.10147. [DOI] [PubMed] [Google Scholar]
- 68.Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–26. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuhn L, Schramm DB, Donninger S, Meddows-Taylor S, Coovadia AH, et al. African infants’ CCL3 gene copies influence perinatal HIV transmission in the absence of maternal nevirapine. AIDS. 2007;21:1753–61. doi: 10.1097/QAD.0b013e3282ba553a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kumar RA, KaraMohamed S, Sudi J, Conrad DF, Brune C, et al. Recurrent 16p11.2 microdeletions in autism. Hum. Mol. Genet. 2008;17:628–38. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 71.Kurotaki N, Shen JJ, Touyama M, Kondoh T, Visser R, et al. Phenotypic consequences of genetic variation at hemizygous alleles: Sotos syndrome is a contiguous gene syndrome incorporating coagulation factor twelve (FXII) deficiency. Genet. Med. 2005;7:479–83. doi: 10.1097/01.gim.0000177419.43309.37. [DOI] [PubMed] [Google Scholar]
- 72.Kurotaki N, Stankiewicz P, Wakui K, Niikawa N, Lupski JR. Sotos syndrome common deletion is mediated by directly oriented subunits within inverted Sos-REP low-copy repeats. Hum. Mol. Genet. 2005;14:535–42. doi: 10.1093/hmg/ddi050. [DOI] [PubMed] [Google Scholar]
- 73.Lalani SR, Thakuria JV, Cox GF, Wang X, Bi W, et al. 20p12.3 microdeletion predisposes to Wolff-Parkinson-White syndrome with variable neurocognitive deficits. J. Med. Genet. 2009;46:168–75. doi: 10.1136/jmg.2008.061002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lalic T, Vossen RH, Coffa J, Schouten JP, Guc-Scekic M, et al. Deletion and duplication screening in the DMD gene using MLPA. Eur. J. Hum. Genet. 2005;13:1231–34. doi: 10.1038/sj.ejhg.5201465. [DOI] [PubMed] [Google Scholar]
- 75.Lam KW, Jeffreys AJ. Processes of copy-number change in human DNA: the dynamics of α-globin gene deletion. Proc. Natl. Acad. Sci. USA. 2006;103:8921–27. doi: 10.1073/pnas.0602690103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lam KW, Jeffreys AJ. Processes of de novo duplication of human alpha-globin genes. Proc. Natl. Acad. Sci. USA. 2007;104:10950–55. doi: 10.1073/pnas.0703856104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Le Marechal C, Masson E, Chen JM, Morel F, Ruszniewski P, et al. Hereditary pancreatitis caused by triplication of the trypsinogen locus. Nat. Genet. 2006;38:1372–74. doi: 10.1038/ng1904. [DOI] [PubMed] [Google Scholar]
- 78.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–47. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 79.Lejeune J, Gautier M, Turpin R. Etude des chromosomes somatiques de neuf enfants mongoliens. C. R. Acad. Sci. 1959;248:1721–22. [PubMed] [Google Scholar]
- 80.Lenski C, Kooy RF, Reyniers E, Loessner D, Wanders RJ, et al. The reduced expression of the HADH2 protein causes X-linked mental retardation, choreoathetosis, and abnormal behavior. Am. J. Hum. Genet. 2007;80:372–77. doi: 10.1086/511527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lieber MR. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 83.Lieber MR, Lu H, Gu J, Schwarz K. Flexibility in the order of action and in the enzymology of the nuclease, polymerases, and ligase of vertebrate nonhomologous DNA end joining: relevance to cancer, aging, and the immune system. Cell Res. 2008;18:125–33. doi: 10.1038/cr.2007.108. [DOI] [PubMed] [Google Scholar]
- 84.Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human nonhomologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 2003;4:712–20. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 85.Lifton RP, Dluhy RG, Powers M, Rich GM, Cook S, et al. A chimeric 11 beta-hydroxylase/aldosterone synthase gene causes glucocorticoid-remediable aldosteronism and human hypertension. Nature. 1992;355:262–65. doi: 10.1038/355262a0. [DOI] [PubMed] [Google Scholar]
- 86.Lindsay SJ, Khajavi M, Lupski JR, Hurles ME. A chromosomal rearrangement hotspot can be identified from population genetic variation and is coincident with a hotspot for allelic recombination. Am. J. Hum. Genet. 2006;79:890–902. doi: 10.1086/508709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lu XY, Phung MT, Shaw CA, Pham K, Neil SE, et al. Genomic imbalances in neonates with birth defects: high detection rates by using chromosomal microarray analysis. Pediatrics. 2008;122:1310–18. doi: 10.1542/peds.2008-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–22. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- 89.Lupski JR. Hotspots of homologous recombination in the human genome: not all homologous sequences are equal. Genome Biol. 2004;5:242. doi: 10.1186/gb-2004-5-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lupski JR. Genome structural variation and sporadic disease traits. Nat. Genet. 2006;38:974–76. doi: 10.1038/ng0906-974. [DOI] [PubMed] [Google Scholar]
- 91.Lupski JR. An evolution revolution provides further revelation. BioEssays. 2007;29:1182–84. doi: 10.1002/bies.20686. [DOI] [PubMed] [Google Scholar]
- 92.Lupski JR. Genomic rearrangements and sporadic disease. Nat. Genet. 2007;39:S43–S7. doi: 10.1038/ng2084. [DOI] [PubMed] [Google Scholar]
- 93.Lupski JR. Structural variation in the human genome. N. Engl. J. Med. 2007;356:1169–71. doi: 10.1056/NEJMcibr067658. [DOI] [PubMed] [Google Scholar]
- 94.Lupski JR. Schizophrenia: incriminating genomic evidence. Nature. 2008;455:178–79. doi: 10.1038/455178a. [DOI] [PubMed] [Google Scholar]
- 95.Lupski JR. Genomic disorders ten years on. Genome Med. 2009;1:42. doi: 10.1186/gm42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lupski JR, Chance PF. Hereditary motor and sensory neuropathies involving altered dosage or mutation of PMP22: the CMT1A duplication and HNPP deletion. In: Dyck PJ, Thomas PK, editors. Peripheral Neuropathy. Elsevier; Philadelphia: 2005. pp. 1659–80. [Google Scholar]
- 97.Lupski JR, de Oca-Luna RM, Slaugenhaupt S, Pentao L, Guzzetta V, et al. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell. 1991;66:219–32. doi: 10.1016/0092-8674(91)90613-4. [DOI] [PubMed] [Google Scholar]
- 98.Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lupski JR, Wise CA, Kuwano A, Pentao L, Parke JT, et al. Gene dosage is a mechanism for Charcot-Marie-Tooth disease type 1A. Nat. Genet. 1992;1:29–33. doi: 10.1038/ng0492-29. [DOI] [PubMed] [Google Scholar]
- 100.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 2008;82:477–88. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Matise TC, Chakravarti A, Patel PI, Lupski JR, Nelis E, et al. Detection of tandem duplications and implications for linkage analysis. Am. J. Hum. Genet. 1994;54:1110–21. [PMC free article] [PubMed] [Google Scholar]
- 102.McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, et al. Common deletion polymorphisms in the human genome. Nat. Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 103.McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat. Genet. 2008;40:1107–12. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 2008;40:1166–74. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 105.Mefford HC, Clauin S, Sharp AJ, Moller RS, Ullmann R, et al. Recurrent reciprocal genomic rearrangements of 17q12 are associated with renal disease, diabetes, and epilepsy. Am. J. Hum. Genet. 2007;81:1057–69. doi: 10.1086/522591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N. Engl. J. Med. 2008;359:1685–99. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Menten P, Wuyts A, Van Damme J. Macrophage inflammatory protein-1. Cytokine Growth Factor Rev. 2002;13:455–81. doi: 10.1016/s1359-6101(02)00045-x. [DOI] [PubMed] [Google Scholar]
- 108.Meyer zu Horste G, Prukop T, Liebetanz D, Mobius W, Nave KA, Sereda MW. Antiprogesterone therapy uncouples axonal loss from demyelination in a transgenic rat model of CMT1A neuropathy. Ann. Neurol. 2007;61:61–72. doi: 10.1002/ana.21026. [DOI] [PubMed] [Google Scholar]
- 109.Miller DW, Hague SM, Clarimon J, Baptista M, Gwinn-Hardy K, et al. Alpha-synuclein in blood and brain from familial Parkinson disease with SNCA locus triplication. Neurology. 2004;62:1835–38. doi: 10.1212/01.wnl.0000127517.33208.f4. [DOI] [PubMed] [Google Scholar]
- 110.Mills RE, Luttig CT, Larkins CE, Beauchamp A, Tsui C, et al. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 2006;16:1182–90. doi: 10.1101/gr.4565806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–23. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Musio A, Selicorni A, Focarelli ML, Gervasini C, Milani D, et al. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat. Genet. 2006;38:528–30. doi: 10.1038/ng1779. [DOI] [PubMed] [Google Scholar]
- 113.Myers S, Freeman C, Auton A, Donnelly P, McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat. Genet. 2008;40:1124–29. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- 114.Myers SR, McCarroll SA. New insights into the biological basis of genomic disorders. Nat. Genet. 2006;38:1363–64. doi: 10.1038/ng1206-1363. [DOI] [PubMed] [Google Scholar]
- 115.Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Naruse K, Ueno M, Satoh T, Nomiyama H, Tei H, et al. A YAC contig of the human CC chemokine genes clustered on chromosome 17q11.2. Genomics. 1996;34:236–40. doi: 10.1006/geno.1996.0274. [DOI] [PubMed] [Google Scholar]
- 117.Nathans J, Piantanida TP, Eddy RL, Shows TB, Hogness DS. Molecular genetics of inherited variation in human color vision. Science. 1986;232:203–10. doi: 10.1126/science.3485310. [DOI] [PubMed] [Google Scholar]
- 118.Nathans J, Thomas D, Hogness DS. Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science. 1986;232:193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- 119.Nguyen DQ, Webber C, Hehir-Kwa J, Pfundt R, Veltman J, Ponting CP. Reduced purifying selection prevails over positive selection in human copy number variant evolution. Genome Res. 2008;18:1711–23. doi: 10.1101/gr.077289.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nobile C, Toffolatti L, Rizzi F, Simionati B, Nigro V, et al. Analysis of 22 deletion breakpoints in dystrophin intron 49. Hum. Genet. 2002;110:418–21. doi: 10.1007/s00439-002-0721-7. [DOI] [PubMed] [Google Scholar]
- 121.Ohno S. Evolution by Gene Duplication. Springer-Verlag; Berlin: 1970. [Google Scholar]
- 122.Ostertag EM, Goodier JL, Zhang Y, Kazazian HH., Jr. SVA elements are nonautonomous retrotransposons that cause disease in humans. Am. J. Hum. Genet. 2003;73:1444–51. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ostertag EM, Kazazian HH., Jr. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 2001;35:501–38. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 124.Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat. Genet. 2007;39:830–32. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Perry GH, Ben-Dor A, Tsalenko A, Sampas N, Rodriguez-Revenga L, et al. The fine-scale and complex architecture of human copy-number variation. Am. J. Hum. Genet. 2008;82:685–95. doi: 10.1016/j.ajhg.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, et al. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 2007;39:1256–60. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pinkel D, Segraves R, Sudar D, Clark S, Poole I, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 1998;20:207–11. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 128.Polymeropoulos MH, Higgins JJ, Golbe LI, Johnson WG, Ide SE, et al. Mapping of a gene for Parkinson’s disease to chromosome 4q21-q23. Science. 1996;274:1197–99. doi: 10.1126/science.274.5290.1197. [DOI] [PubMed] [Google Scholar]
- 129.Popesco MC, Maclaren EJ, Hopkins J, Dumas L, Cox M, et al. Human lineage-specific amplification, selection, and neuronal expression of DUF1220 domains. Science. 2006;313:1304–7. doi: 10.1126/science.1127980. [DOI] [PubMed] [Google Scholar]
- 130.Potocki L, Bi W, Treadwell-Deering D, Carvalho CM, Eifert A, et al. Characterization of Potocki-Lupski syndrome (dup(17)(p11.2p11.2)) and delineation of a dosage-sensitive critical interval that can convey an autism phenotype. Am. J. Hum. Genet. 2007;80:633–49. doi: 10.1086/512864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Raedt TD, Stephens M, Heyns I, Brems H, Thijs D, et al. Conservation of hotspots for recombination in low-copy repeats associated with the NF1 microdeletion. Nat. Genet. 2006;38:1419–23. doi: 10.1038/ng1920. [DOI] [PubMed] [Google Scholar]
- 132.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rovelet-Lecrux A, Hannequin D, Raux G, Le Meur N, Laquerriere A, et al. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat. Genet. 2006;38:24–26. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- 134.Sahin-Toth M. Biochemical models of hereditary pancreatitis. Endocrinol. Metab. Clin. North Am. 2006;35:303–12. ix. doi: 10.1016/j.ecl.2006.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Saunier S, Calado J, Benessy F, Silbermann F, Heilig R, et al. Characterization of the NPHP1 locus: mutational mechanism involved in deletions in familial juvenile nephronophthisis. Am. J. Hum. Genet. 2000;66:778–89. doi: 10.1086/302819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schwarz K, Ma Y, Pannicke U, Lieber MR. Human severe combined immune deficiency and DNA repair. BioEssays. 2003;25:1061–70. doi: 10.1002/bies.10344. [DOI] [PubMed] [Google Scholar]
- 137.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–49. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–28. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 139.Sen SK, Han K, Wang J, Lee J, Wang H, et al. Human genomic deletions mediated by recombination between Alu elements. Am. J. Hum. Genet. 2006;79:41–53. doi: 10.1086/504600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sennvik K, Fastbom J, Blomberg M, Wahlund LO, Winblad B, Benedikz E. Levels of alpha- and beta-secretase cleaved amyloid precursor protein in the cerebrospinal fluid of Alzheimer’s disease patients. Neurosci. Lett. 2000;278:169–72. doi: 10.1016/s0304-3940(99)00929-5. [DOI] [PubMed] [Google Scholar]
- 141.Sereda MW, Meyer zu Horste G, Suter U, Uzma N, Nave KA. Therapeutic administration of progesterone antagonist in a model of Charcot-Marie-Tooth disease (CMT-1A) Nat. Med. 2003;9:1533–37. doi: 10.1038/nm957. [DOI] [PubMed] [Google Scholar]
- 142.Shaffer LG, Lupski JR. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu. Rev. Genet. 2000;34:297–329. doi: 10.1146/annurev.genet.34.1.297. [DOI] [PubMed] [Google Scholar]
- 143.Sharp AJ, Hansen S, Selzer RR, Cheng Z, Regan R, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat. Genet. 2006;38:1038–42. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 144.Sharp AJ, Mefford HC, Li K, Baker C, Skinner C, et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat. Genet. 2008;40:322–28. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sharp AJ, Selzer RR, Veltman JA, Gimelli S, Gimelli G, et al. Characterization of a recurrent 15q24 microdeletion syndrome. Hum. Mol. Genet. 2007;16:567–72. doi: 10.1093/hmg/ddm016. [DOI] [PubMed] [Google Scholar]
- 146.Shaw-Smith C, Redon R, Rickman L, Rio M, Willatt L, et al. Microarray based comparative genomic hybridisation (array-CGH) detects submicroscopic chromosomal deletions and duplications in patients with learning disability/mental retardation and dysmorphic features. J. Med. Genet. 2004;41:241–48. doi: 10.1136/jmg.2003.017731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shaw CJ, Lupski JR. Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum. Mol. Genet. 2004;13(Spec No 1):R57–64. doi: 10.1093/hmg/ddh073. [DOI] [PubMed] [Google Scholar]
- 148.Shaw CJ, Lupski JR. Non-recurrent 17p11.2 deletions are generated by homologous and nonhomologous mechanisms. Hum. Genet. 2005;116:1–7. doi: 10.1007/s00439-004-1204-9. [DOI] [PubMed] [Google Scholar]
- 149.She X, Cheng Z, Zollner S, Church DM, Eichler EE. Mouse segmental duplication and copy number variation. Nat. Genet. 2008;40:909–14. doi: 10.1038/ng.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, et al. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 151.Sleegers K, Brouwers N, Gijselinck I, Theuns J, Goossens D, et al. APP duplication is sufficient to cause early onset Alzheimer’s dementia with cerebral amyloid angiopathy. Brain. 2006;129:2977–83. doi: 10.1093/brain/awl203. [DOI] [PubMed] [Google Scholar]
- 152.Smith GR. Chi sites and their consequences. In: de Bruijn FJ, Lupski JR, Weinstock GM, editors. Bacterial Genomes: Physical Structure and Analysis. Chapman & Hall; New York: 1998. pp. 49–66. [Google Scholar]
- 153.Stankiewicz P, Beaudet AL. Use of array CGH in the evaluation of dysmorphology, malformations, developmental delay, and idiopathic mental retardation. Curr. Opin. Genet. Dev. 2007;17:182–92. doi: 10.1016/j.gde.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 154.Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 155.Stankiewicz P, Shaw CJ, Dapper JD, Wakui K, Shaffer LG, et al. Genome architecture catalyzes nonrecurrent chromosomal rearrangements. Am. J. Hum. Genet. 2003;72:1101–16. doi: 10.1086/374385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–36. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Steinmann K, Cooper DN, Kluwe L, Chuzhanova NA, Senger C, et al. Type 2 NF1 deletions are highly unusual by virtue of the absence of nonallelic homologous recombination hotspots and an apparent preference for female mitotic recombination. Am. J. Hum. Genet. 2007;81:1201–20. doi: 10.1086/522089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–53. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.The Int. HapMap Consort. The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 160.The Int. HapMap Consort. A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.The Int. Schizophrenia Consort. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–41. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Theuns J, Brouwers N, Engelborghs S, Sleegers K, Bogaerts V, et al. Promoter mutations that increase amyloid precursor-protein expression are associated with Alzheimer disease. Am. J. Hum. Genet. 2006;78:936–46. doi: 10.1086/504044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tiemann-Boege I, Calabrese P, Cochran DM, Sokol R, Arnheim N. High-resolution recombination patterns in a region of human chromosome 21 measured by sperm typing. PLoS Genet. 2006;2:e70. doi: 10.1371/journal.pgen.0020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Toffolatti L, Cardazzo B, Nobile C, Danieli GA, Gualandi F, et al. Investigating the mechanism of chromosomal deletion: characterization of 39 deletion breakpoints in introns 47 and 48 of the human dystrophin gene. Genomics. 2002;80:523–30. [PubMed] [Google Scholar]
- 165.Townson JR, Barcellos LF, Nibbs RJ. Gene copy number regulates the production of the human chemokine CCL3-L1. Eur. J. Immunol. 2002;32:3016–26. doi: 10.1002/1521-4141(2002010)32:10<3016::AID-IMMU3016>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 166.Turner DJ, Miretti M, Rajan D, Fiegler H, Carter NP, et al. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat. Genet. 2008;40:90–95. doi: 10.1038/ng.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, et al. Fine-scale structural variation of the human genome. Nat. Genet. 2005;37:727–32. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 168.Van Esch H, Bauters M, Ignatius J, Jansen M, Raynaud M, et al. Duplication of the MECP2 region is a frequent cause of severe mental retardation and progressive neurological symptoms in males. Am. J. Hum. Genet. 2005;77:442–53. doi: 10.1086/444549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.van Ommen GJ. Frequency of new copy number variation in humans. Nat. Genet. 2005;37:333–34. doi: 10.1038/ng0405-333. [DOI] [PubMed] [Google Scholar]
- 170.Velagaleti GV, Bien-Willner GA, Northup JK, Lockhart LH, Hawkins JC, et al. Position effects due to chromosome breakpoints that map approximately 900 Kb upstream and approximately 1.3 Mb downstream of SOX9 in two patients with campomelic dysplasia. Am. J. Hum. Genet. 2005;76:652–62. doi: 10.1086/429252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Vissers LE, Stankiewicz P, Yatsenko SA, Crawford E, Creswick H, et al. Complex chromosome 17p rearrangements associated with low-copy repeats in two patients with congenital anomalies. Hum. Genet. 2007;121:697–709. doi: 10.1007/s00439-007-0359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Vrijenhoek T, Buizer-Voskamp JE, Van Der Stelt I, Strengman E, Genetic Risk and Outcome in Psychosis (GROUP) Consort. et al. Recurrent CNVs disrupt three candidate genes in schizophrenia patients. Am. J. Hum. Genet. 2008;83:504–10. doi: 10.1016/j.ajhg.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–43. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 174.Wang J, Wang W, Li R, Li Y, Tian G, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 2008;358:667–75. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 176.Wellcome WTCCC Genome-wide association study of 14000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–76. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 178.Whitcomb DC, Gorry MC, Preston RA, Furey W, Sossenheimer MJ, et al. Hereditary pancreatitis is caused by a mutation in the cationic trypsinogen gene. Nat. Genet. 1996;14:141–45. doi: 10.1038/ng1096-141. [DOI] [PubMed] [Google Scholar]
- 179.Willcocks LC, Lyons PA, Clatworthy MR, Robinson JI, Yang W, et al. Copy number of FCGR3B, which is associated with systemic lupus erythematosus, correlates with protein expression and immune complex uptake. J. Exp. Med. 2008;205:1573–82. doi: 10.1084/jem.20072413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wolf NI, Sistermans EA, Cundall M, Hobson GM, Davis-Williams AP, et al. Three or more copies of the proteolipid protein gene PLP1 cause severe Pelizaeus-Merzbacher disease. Brain. 2005;128:743–51. doi: 10.1093/brain/awh409. [DOI] [PubMed] [Google Scholar]
- 181.Xing J, Zhang Y, Han K, Salem AH, Sen SK, et al. Mobile elements create structural variation: analysis of a complete human genome. Genome Res. 2009 doi: 10.1101/gr.091827.109. In press. doi:10.1101/gr.091827.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat. Genet. 2008;40:880–85. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 183.Yan J, Bi W, Lupski JR. Penetrance of craniofacial anomalies in mouse models of Smith-Magenis syndrome is modified by genomic sequence surrounding Rai1: Not all null alleles are alike. Am. J. Hum. Genet. 2007;80:518–25. doi: 10.1086/512043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yang Y, Chung EK, Wu YL, Savelli SL, Nagaraja HN, et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): Low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am. J. Hum. Genet. 2007;80:1037–54. doi: 10.1086/518257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhang F, Carvalho CM, Lupski JR. Complex human chromosomal and genomic rearrangements. Trends Genet. 2009 doi: 10.1016/j.tig.2009.05.005. In press, doi: 10.1016/j.tig.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zhang F, Khajavi M, Connolly AM, Towne CF, Batish SD, Lupski JR. The DNA replication FoSTeS/MMBIR mechanism can generate human genomic, genic, and exonic complex rearrangements. Nat. Genet. 2009;41:849–53. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.