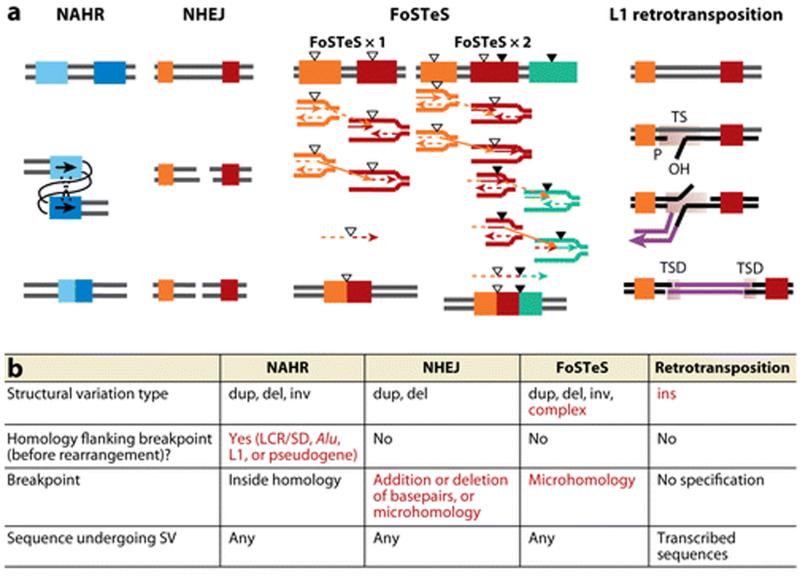
Figure 3.
Comparisons and characteristics of the four major mechanisms underlying human genomic rearrangements and CNV formation. (a) Models for Non-Allelic Homologous Recombination (NAHR) between repeat sequences (LCRs/SDs, Alu, or L1 elements); Non-Homologous End-Joining (NHEJ), recombination repair of double strand break; Fork Stalling and Template Switching (FoSTeS), multiple FoSTeS events (×2 or more) resulting in complex rearrangement and single FoSTeS event (×1) causing simple rearrangement; and retrotransposition. TS, target site; TSD, duplicated target site. Adapted from References (47, 123). Thick bars of different colors indicate different genomic fragments; completely different colors (as orange and red or orange/red/green in FoSTeS×2) symbolize that no homology between the two fragments is required. The two bars in two similar shades of blue indicate that the two fragments involved in NAHR should have extensive homology with each other. The triangles symbolize short sequences sharing microhomologies. Each group of triangles (either filled or empty) indicates one group of sequences sharing the same microhomology with each other. (b) Characteristic features for each rearrangement mechanism. Specific features of certain mechanisms are shown in red. Abbreviations: dup, duplication; del, deletion; inv, inversion; ins, insertion.