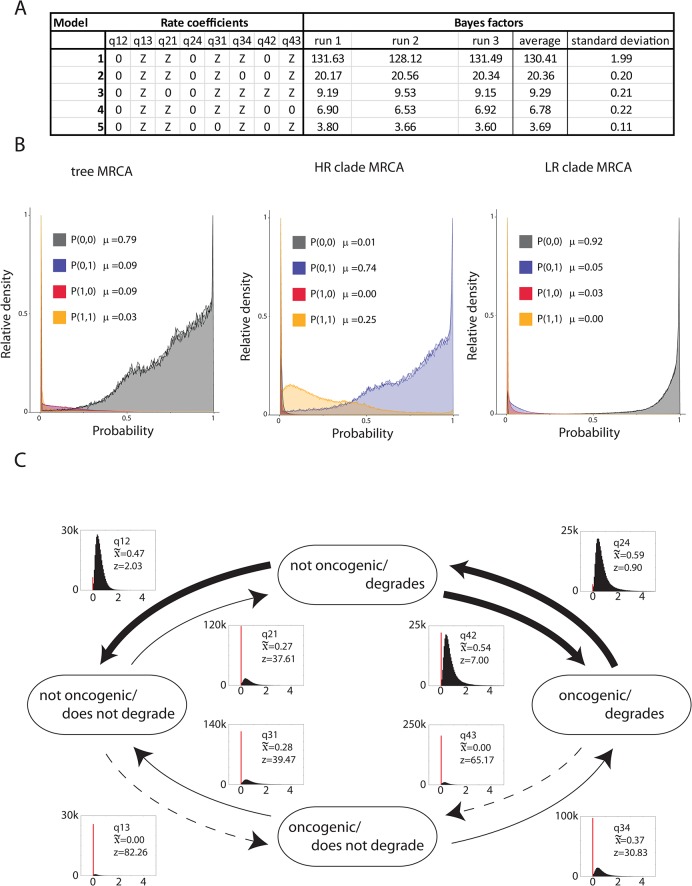
Fig 3. Degradation of PDZ proteins is an enabling phenotype towards oncogenicity.
(A) Table of top five models as selected by the RJMCMC analysis. The RJMCMC was run three independent times to ensure that the reproducibility of the model analysis. Through comparing the ratio of posterior to prior odds (see materials and methods) we obtain Bayes Factors to support the choice of a specific model. The selected model has a Bayes Factor of 130.41 (st.dev. = 1.99) suggesting decisive evidence in favor of this model. (B) Ancestral phenotype reconstruction at three important nodes of the Alphapapillomavirus phylogeny. The graphs show the estimated marginal probability density plot for the common MRCA, the high-risk MRCA or the low-risk MRCA. The analysis suggests that the MRCA of the high-risk viruses acquired the ability to degrade PDZ-containing protein, but was likely not an oncogenic virus. (C) Estimated instantaneous rates of change between different combinations of viral phenotypes based on the RJMCMC analysis. Histograms show the posterior distribution of estimated values of the rate parameters. The red bars indicate the fraction of samples in which each rate is estimated to be zero. Arrow width is proportional to the median of these estimated rates. Z indicates the percentage of samples in which each rate parameter is estimated as zero. Consistent with the hypothesis that the ability to degrade PDZ containing proteins is an enabling phenotype, rates associated with oncogenic ability independent of PDZ protein interaction are often estimated as zero (i.e. they do not occur), and are thus represented as dotted line.