Abstract
Fragaria × ananassa (common name: strawberry) is a globally cultivated hybrid species belonging to Rosaceae family. Colletotrichum acutatum sensu lato (s.l.) is considered to be the second most economically important pathogen worldwide affecting strawberries. A collection of 148 Colletotrichum spp. isolates including 67 C. acutatum s.l. isolates associated with the phytosanitary history of UK strawberry production were used to characterize multi-locus genetic variation of this pathogen in the UK, relative to additional reference isolates that represent a worldwide sampling of the diversity of the fungus. The evidence indicates that three different species C. nymphaeae, C. godetiae and C. fioriniae are associated with strawberry production in the UK, which correspond to previously designated genetic groups A2, A4 and A3, respectively. Among these species, 12 distinct haplotypes were identified suggesting multiple introductions into the country. A subset of isolates was also used to compare aggressiveness in causing disease on strawberry plants and fruits. Isolates belonging to C. nymphaeae, C. godetiae and C. fioriniae representative of the UK anthracnose pathogen populations showed variation in their aggressiveness. Among the three species, C. nymphaeae and C. fioriniae appeared to be more aggressive compared to C. godetiae. This study highlights the genetic and pathogenic heterogeneity of the C. acutatum s.l. populations introduced into the UK linked to strawberry production.
Introduction
Fragaria × ananassa (common name: strawberry) is a hybrid species cultivated worldwide belonging to the Rosaceae family. Since the 1980s, the UK strawberry industry has expanded rapidly representing a significant component of fruit production in the country [1]. Anthracnose is a major disease of cultivated strawberry, caused by two species complexes of the fungus referred to as C. acutatum and C. gloeosporioides. C. acutatum is considered to be the dominant cause of strawberry anthracnose, and the second most important pathogen of strawberry after Botrytis cinerea [2–7]. The C. gloeosporioides complex includes C. fragariae, which is now considered synonymous with a new species C. theobromicola [8]. However, researchers have often continued to use the name C. fragariae when referring to a pathogen that was associated with strawberry anthracnose [9–12]. C. gloeosporioides is found only occasionally on strawberry in Europe [3,7].
C. acutatum s.l. was described for the first time as a strawberry pathogen in California in 1983 [13], and has since appeared to have spread worldwide, including the UK, through runners and propagating material [2,6,14–16]. A first extensive genetic characterization of C. acutatum s.l. representing the global diversity of the pathogen led to its sub-division into genetic groups named from A1 to A9 [6, 17]. More recently, the C. acutatum s.l. has been sub-divided into more than 30 species based on multi-locus phylogeny [18].
The first record of C. acutatum s.l. in the UK was in 1978, on Anemone sp. grown in Jersey [19]. In 1982, the first incidence of anthracnose disease in strawberries caused by C. acutatum s.l. was recorded in the UK, and was attributed to the importation of infected strawberry runners from the USA [20]. DNA sequences in public databases suggest two UK isolates (CBS198.35 and CBS199.35) that were collected in 1935 from the host Phormium spp. (common name “New Zealand flax”) belong to C. acutatum s.l. [18, 21]. CABI database records during 1978 to 1983 shows the incidence of the pathogens various hosts and in different locations in the UK (http://www.herbimi.info). However, it seems highly improbable that the first outbreak on strawberry led to the wide dispersal of the pathogen. In 1993, Lovelidge proposed that the continued introduction of infected strawberry material from abroad was so common that the disease was destined to become endemic in the UK [14]. In subsequent years, further outbreaks have been reported on strawberry linked to the importation of infected propagation material mainly from mainland Europe and on other important crop hosts [20,22,23].
Strawberry anthracnose symptoms produced by the two Colletotrichum species complexes are similar and can be found on all parts of the plant [12]. Flower blight and fruit rot are common symptoms in the field [24], whereas lesions on stolons, petioles and leaves are mainly found in plant nurseries [15]. Crown symptomatology is characterized by reddish-brown necrotic areas [25] and in some cases stunting and chlorosis have been associated with root necrosis [15].
Research has been carried out to characterize C. acutatum s.l. populations related to strawberry in specific geographic areas including Israel, France, Bulgaria, Spain, Belgium and other European countries [2–5,7,26] and from specific regions of the USA [25]. Other research has attempted to characterize C. acutatum s.l. related to strawberry using isolates collected worldwide [3], both by genomic fingerprinting (such as RFLP, apPCR, etc.) and sequence analysis based on the ITS region. Results have highlighted the presence of at least one representative “clonal” population suggesting a single source of origin and, consequentially, that the disease is spread through infected propagation material. However, ITS sequences alone or genomic fingerprinting are not suitable to discriminate among the newly assigned species designations.
In a recent study based on the analysis of more than two decades of anthracnose incidence data sets gathered by authorities responsible for plant health, trade was identified as the main route of entry and establishment of C. acutatum in the UK strawberry production. Over this period, various nurseries were importing planting material into the UK, and at least 55 cases of infested material that was planted in the field through imports that were not intercepted by the border inspection posts, were identified [20].
The focus of the present study was to assess the extent of the genetic and pathogenic diversity of these introduced pathogen populations mainly utilising a unique collection of C. acutatum s. l. isolates established through the plant health inspection surveys from the early 1980s onwards. We focused on C. acutatum s.l. because previous reports from France, Israel, UK, Bulgaria and Spain had described this taxa as a major widely distributed pathogen, compared with other species such as C. gloeosporioides s.l. that occur less frequently in Europe [2–5,12]. A range of historic and contemporary C. acutatum s. l. isolates including those from worldwide strawberry crops, other plant hosts in the UK, as well as worldwide representatives from different hosts building on our previous work were accessed as reference sources for determining the genetic and species identities of isolates associated with UK strawberry anthracnose phytosanitary control work. Based on multi-locus phylogenetic analysis, we have identified 12 different haplotypes that belong to three different species C. nymphaeae, C. godetiae and C. fioriniae suggesting multiple introductions of the strawberry anthracnose pathogen. Pathogenic and growth characteristics of these haplotype representatives further highlight the heterogeneity of the introduced pathogen populations.
Materials and Methods
Fungal isolates and culture conditions
A diverse collection of C. acutatum s.l. was assembled for this study including: 67 isolates associated with strawberry production in the UK (obtained from the UK Food and Environment Research Agency, or FERA responsible for plant health within the Department for Environment, Food and Rural Affairs), 27 C. acutatum s.l. isolates collected from strawberry in other countries, and 13 isolates collected from other host species in the UK. For further comparison, 33 isolates were added to represent other genetic groups, and novel species from previous studies [6,17,18]. This included two isolates of C. fruticola, two isolates of C. aenigma (belonging to C. gloeosporioides species complex [8]) associated with strawberry, two UK isolates of C. spinaciae and one isolate each of C. graminicola, C. higginsianum [27] and C. fioriniae [28]. Sequence data of the markers was retrieved from the reference genome sequences available from Genbank for C. graminicola and C. higginsianum (accession numbers: ACOD01000000 and CACQ02000000, respectively) used among out-groups in the phylogenetic analysis (Fig 1). Details of the isolate collection used in the present study are provided in Table 1.
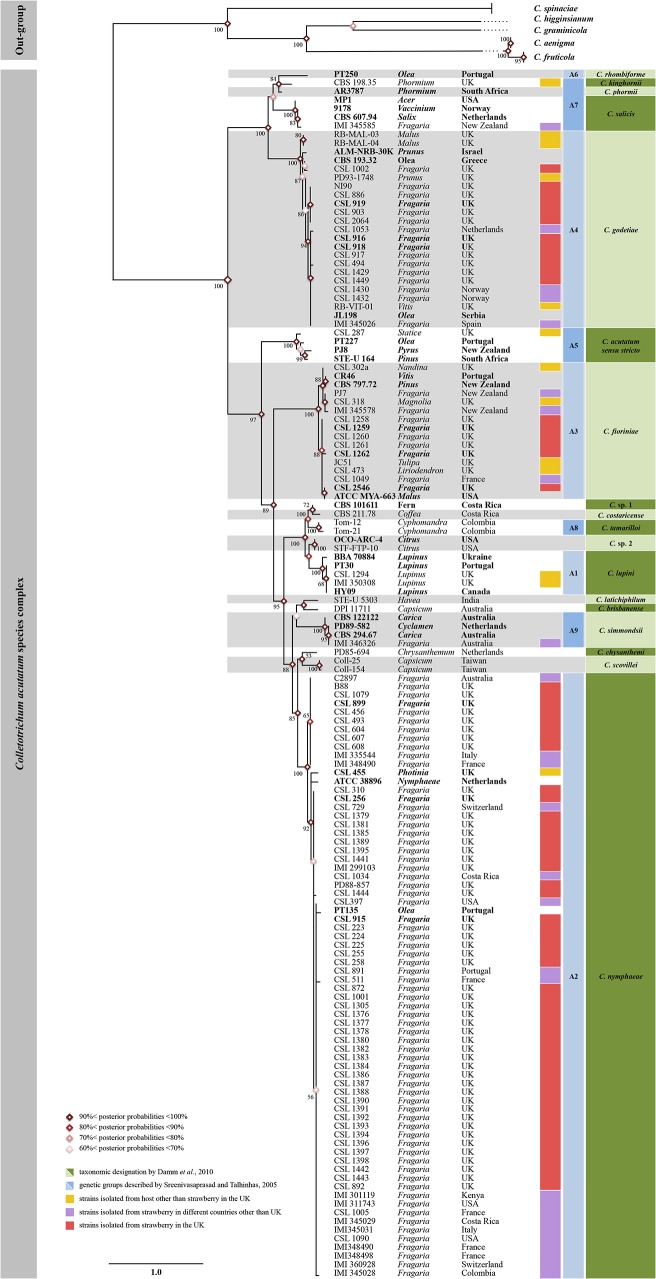
Fig 1. Multilocus phylogenetic analysis of the Colletotrichum isolates used in this study.
Bayesian MCMC analysis tree constructed from the alignment based on the concatenation of rRNA, TUB, MAT1-2 and GPDH partial sequences of 140 Colletotrichum acutatum sensu lato isolates used in this study. The tree was rooted with sequences from C. graminicola and C. higginsianum retrieved from whole genome sequences and sequences of four C. gloeosporioides sensu lato and two C. spinaciae obtained experimentally. Isolates used to investigate variation in aggressiveness are highlighted in bold.
Table 1. Colletotrichum sp. strains used in this study with isolation details and GenBank accessions.
| Strain Code | Genus | Species | Genetic group [6] | Country | Host | Accession numbers | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| ITS | TUB | MAT1-2 | GAPDH | |||||||
| Isolates from strawberry in UK | ||||||||||
| B88 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246514 | KM251867 | KM251969 | KM252115 | |
| NI90 | Colletotrichum | godetiae | A4 | United Kingdom | Fragaria x ananassa | AF411766 | AJ409294 | KM251970 | KM252116 | |
| CSL 1079 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246515 | KM251868 | KM251981 | KM252118 | |
| CSL 2546 | Colletotrichum | fioriniae | A3 | United Kingdom | Fragaria x ananassa | KM246516 | KM251870 | KM251983 | KM252120* | |
| CSL 899 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246518 | KM251872 | KM251985 | KM252122* | |
| CSL 310 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246519 | KM251873 | KM251986 | KM252123 | |
| CSL 915 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246520 | KM251874 | KM251987 | KM252124* | |
| CSL 886 | Colletotrichum | godetiae | A4 | United Kingdom | Fragaria x ananassa | KM246521 | KM251875 | KM251988 | KM252125 | |
| CSL 919 | Colletotrichum | godetiae | A4 | United Kingdom | Fragaria x ananassa | KM246522 | KM251876 | KM251989 | KM252126* | |
| CSL 916 | Colletotrichum | godetiae | A4 | United Kingdom | Fragaria x ananassa | KM246523 | KM251877 | KM251990 | KM252127* | |
| CSL 918 | Colletotrichum | godetiae | A4 | United Kingdom | Fragaria x ananassa | KM246524 | KM251878 | KM251991 | KM252128* | |
| CSL 917 | Colletotrichum | godetiae | A4 | United Kingdom | Fragaria x ananassa | KM246525 | KM251879 | KM251992 | KM252129 | |
| CSL 223 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246526 | KM251880 | KM251993 | KM252130 | |
| CSL 224 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246527 | KM251881 | KM251994 | KM252131 | |
| CSL 225 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246528 | KM251882 | KM251995 | KM252132 | |
| CSL 255 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246529 | KM251883 | KM251996 | KM252133 | |
| CSL 256 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246530 | KM251884 | KM251997 | KM252134* | |
| CSL 258 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246531 | KM251885 | KM251998 | KM252135 | |
| CSL 456 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria vesca | KM246532 | KM251886 | KM251999 | KM252136 | |
| CSL 493 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246533 | KM251887 | KM252000 | KM252137 | |
| CSL 494 | Colletotrichum | godetiae | A4 | United Kingdom | Fragaria vesca | KM246534 | KM251888 | KM252001 | KM252138 | |
| CSL 604 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246535 | KM251890 | KM252003 | KM252140 | |
| CSL 607 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246538 | KM251893 | KM252006 | KM252143 | |
| CSL 608 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246539 | KM251894 | KM252007 | KM252144 | |
| CSL 872 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246541 | KM251896 | KM252009 | KM252146 | |
| CSL 903 | Colletotrichum | godetiae | A4 | United Kingdom | Fragaria x ananassa | KM246542 | KM251897 | KM252010 | KM252147 | |
| CSL 1001 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246543 | KM251898 | KM252011 | KM252148 | |
| CSL 1258 | Colletotrichum | fioriniae | A3 | United Kingdom | Fragaria x ananassa | KM246544 | KM251899 | KM252012 | KM252149 | |
| CSL 1259 | Colletotrichum | fioriniae | A3 | United Kingdom | Fragaria x ananassa | KM246545 | KM251900 | KM252013 | KM252150* | |
| CSL 1260 | Colletotrichum | fioriniae | A3 | United Kingdom | Fragaria x ananassa | KM246546 | KM251901 | KM252014 | KM252151 | |
| CSL 1261 | Colletotrichum | fioriniae | A3 | United Kingdom | Fragaria x ananassa | KM246547 | KM251902 | KM252015 | KM252152 | |
| CSL 1262 | Colletotrichum | fioriniae | A3 | United Kingdom | Fragaria x ananassa | KM246548 | KM251903 | KM252016 | KM252153* | |
| CSL 1305 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246549 | KM251904 | KM252017 | KM252154 | |
| CSL 1376 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246550 | KM251905 | KM252018 | KM252155 | |
| CSL 1377 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246551 | KM251906 | KM252019 | KM252156 | |
| CSL 1378 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246552 | KM251907 | KM252020 | KM252157 | |
| CSL 1379 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246553 | KM251908 | KM252021 | KM252158 | |
| CSL 1380 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246554 | KM251909 | KM252022 | KM252159 | |
| CSL 1381 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246555 | KM251910 | KM252023 | KM252160 | |
| CSL 1382 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246556 | KM251911 | KM252024 | KM252161 | |
| CSL 1383 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246557 | KM251912 | KM252025 | KM252162 | |
| CSL 1384 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246558 | KM251913 | KM252026 | KM252163 | |
| CSL 1385 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246559 | KM251914 | KM252027 | KM252164 | |
| CSL 1386 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246560 | KM251915 | KM252028 | KM252165 | |
| CSL 1387 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246561 | KM251916 | KM252029 | KM252166 | |
| CSL 1388 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246562 | KM251917 | KM252030 | KM252167 | |
| CSL 1389 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246563 | KM251918 | KM252031 | KM252168 | |
| CSL 1390 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246564 | KM251919 | KM252032 | KM252169 | |
| CSL 1391 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246565 | KM251920 | KM252033 | KM252170 | |
| CSL 1392 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246566 | KM251921 | KM252034 | KM252171 | |
| CSL 1393 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246567 | KM251922 | KM252035 | KM252172 | |
| CSL 1394 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246568 | KM251923 | KM252036 | KM252173 | |
| CSL 1395 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246569 | KM251924 | KM252037 | KM252174 | |
| CSL 1396 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246570 | KM251925 | KM252038 | KM252175 | |
| CSL 1397 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246571 | KM251926 | KM252039 | KM252176 | |
| CSL 1398 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246572 | KM251927 | KM252040 | KM252177 | |
| CSL 1429 | Colletotrichum | godetiae | A4 | United Kingdom | Fragaria x ananassa | KM246573 | KM251928 | KM252041 | KM252178 | |
| CSL 1441 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246574 | KM251929 | KM252042 | KM252179 | |
| CSL 1442 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246575 | KM251930 | KM252043 | KM252180 | |
| CSL 1443 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246576 | KM251931 | KM252044 | KM252181 | |
| CSL 1444 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246577 | KM251932 | KM252045 | KM252182 | |
| CSL 1449 | Colletotrichum | godetiae | A4 | United Kingdom | Fragaria x ananassa | KM246578 | KM251933 | KM252046 | KM252183 | |
| CSL 2064 | Colletotrichum | godetiae | A4 | United Kingdom | Fragaria x ananassa | KM246579 | KM251934 | KM252047 | KM252184 | |
| CSL 1002 | Colletotrichum | godetiae | A4 | United Kingdom | Fragaria x ananassa | KM246580 | KM251935 | KM252048 | KM252185 | |
| CSL 892 | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | KM246584 | KM251938 | KM252053 | KM252188 | |
| IMI 299103 [18] | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria vesca | JQ948231 | JQ949882 | KM252069 | JQ948561 | |
| PD88-857, CBS 125973 [18] | Colletotrichum | nymphaeae | A2 | United Kingdom | Fragaria x ananassa | JQ948232 | JQ949883 | KM252100 | JQ948562 | |
| C. acutatum sensu lato from strawberry worldwide | ||||||||||
| C2897 | Colletotrichum | nymphaeae | A2 | Australia | Fragaria x ananassa | AJ300558 | AJ314718 | KM251967 | KM252113 | |
| CSL 397 | Colletotrichum | nymphaeae | A2 | USA | Fragaria x ananassa | AF411765 | AJ409296 | KM251968 | KM252114 | |
| CSL 1053 | Colletotrichum | godetiae | A4 | Netherlands | Fragaria x ananassa | AJ536210 | KM251869 | KM251982 | KM252119 | |
| CSL 891 | Colletotrichum | nymphaeae | A2 | Portugal | Fragaria sp. | EF622184 | KM251889 | KM252002 | KM252139 | |
| CSL 511 | Colletotrichum | nymphaeae | A2 | France | Fragaria x ananassa | KM246536 | KM251891 | KM252004 | KM252141 | |
| CSL 729 | Colletotrichum | nymphaeae | A2 | Switzerland | Fragaria x ananassa | KM246537 | KM251892 | KM252005 | KM252142 | |
| CSL 1430 | Colletotrichum | godetiae | A4 | Norway | Fragaria vesca | KM246585 | KM251939 | KM252054 | KM252189 | |
| CSL 1432 | Colletotrichum | godetiae | A4 | Norway | Fragaria x ananassa | KM246586 | KM251940 | KM252055 | KM252190 | |
| PJ7 [28] | Colletotrichum | fioriniae | A3 | New Zealand | Fragaria x ananassa | genome: JARH00000000 | ||||
| CSL 1020, IMI 301119 [18] | Colletotrichum | nymphaeae | A2 | Kenya | Fragaria vesca | JQ948266 | JQ949917 | KM252070 | JQ948596 | |
| IMI 311743 [18] | Colletotrichum | nymphaeae | A2 | USA | Fragaria x ananassa | JQ948258 | JQ949909 | KM252071 | JQ948588 | |
| IMI 335544 | Colletotrichum | nymphaeae | A2 | Italy | Fragaria x ananassa | KJ018636 | KJ018648 | KM252072 | KJ018660 | |
| IMI 345026 [18] | Colletotrichum | godetiae | A4 | Spain | Fragaria x ananassa | JQ948424 | JQ950075 | KM252073 | JQ948755 | |
| CSL 1005, IMI 345027 | Colletotrichum | nymphaeae | A2 | France | Fragaria x ananassa | AJ536199 | KM251946 | KM252074 | KM252198 | |
| IMI 345028 | Colletotrichum | nymphaeae | A2 | Colombia | Fragaria x ananassa | AF090853 | KM251947 | KM252075 | KM252199 | |
| IMI 345029 | Colletotrichum | nymphaeae | A2 | Costa Rica | Fragaria x ananassa | KM246591 | KM251948 | KM252076 | KM252200 | |
| CSL 1034, IMI345030 | Colletotrichum | nymphaeae | A2 | Costa Rica | Fragaria x ananassa | AJ536203 | KM251949 | KM252077 | KM252201 | |
| IMI 345031 | Colletotrichum | nymphaeae | A2 | Italy | Fragaria x ananassa | KM246592 | KM251950 | KM252078 | KM252202 | |
| IMI 345578 [18] | Colletotrichum | fioriniae | A3 | New Zealand | Fragaria ananassa | JQ948334 | JQ949985 | KM252080 | JQ948664 | |
| CSL 1046, IMI 346326 | Colletotrichum | simmondsii | A2 | Australia | Fragaria x ananassa | AJ536208 | KM251952 | KM252081 | KM252204 | |
| IMI 345585 [18] | Colletotrichum | salicis | A7 | New Zealand | Fragaria x ananassa | JQ948476 | JQ950127 | KM252084 | JQ948807 | |
| CSL 1090, IMI 348160 | Colletotrichum | nymphaeae | A2 | USA | Fragaria x ananassa | AJ536200 | KM251953 | KM252086 | KM252205 | |
| IMI 348177 [18] | Colletotrichum | nymphaeae | A2 | USA | Fragaria x ananassa | KM246593 | KM251954 | KM252087 | KM252206 | |
| IMI 348490 | Colletotrichum | nymphaeae | A2 | France | Fragaria x ananassa | KM246594 | KM251955 | KM252088 | KM252207 | |
| CSL 1086, IMI 348498 | Colletotrichum | nymphaeae | A2 | France | Fragaria x ananassa | KM246595 | KM251956 | KM252089 | KM252208 | |
| CSL 1049, IMI 348499 | Colletotrichum | fioriniae | A3 | France | Fragaria x ananassa | AJ536220 | KM251957 | KM252090 | KM252209 | |
| IMI 360928 [18] | Colletotrichum | nymphaeae | A2 | Switzerland | Fragaria x ananassa | JQ948243 | JQ949894 | KM252091 | JQ948573 | |
| Strains isolated from different hosts in UK | ||||||||||
| RB-MAL-03 [23] | Colletotrichum | godetiae | A4 | United Kingdom | Malus domestica | KF834206 | KF834207 | KM252049 | KF834208 | |
| RB-MAL-04 | Colletotrichum | godetiae | A4 | United Kingdom | Malus domestica | KM246582 | KM251936 | KM252050 | KM252186 | |
| CSL 1294 | Colletotrichum | lupini | A1 | United Kingdom | Lupinus polyphyllus | AJ300561 | KM251944 | KM252059 | KM252194 | |
| CSL 287 [18] | Colletotrichum | acutatum | A5 | United Kingdom | Statice sp. | JQ948389 | JQ950040 | KM252060 | JQ948720 | |
| RB-VIT-01,CBS 129951 [22] | Colletotrichum | godetiae | A4 | United Kingdom | Vitis vinifera | KF834203 | KF834204 | KM252061 | KF834205 | |
| CSL 455 [18] | Colletotrichum | nymphaeae | A2 | United Kingdom | Photinia sp. | JQ948217 | JQ949868 | KM252063 | JQ948547 | |
| JC51, CBS 129948 [18] | Colletotrichum | fioriniae | A3 | United Kingdom | Tulipa sp. | AJ749680 | KM251945 | KM252064 | KM252195 | |
| CSL 302a | Colletotrichum | fioriniae | A3 | United Kingdom | Nandina domestica | AJ749670 | AJ748626 | KM252065 | KM252196 | |
| CSL 473 [18] | Colletotrichum | fioriniae | A3 | United Kingdom | Liriodendron tulipifera | JQ948345 | JQ949996 | KM252066 | JQ948675 | |
| CSL 318 [18] | Colletotrichum | fioriniae | A3 | United Kingdom | Magnolia sp. | JQ948346 | JQ949997 | KM252067 | JQ948676 | |
| IMI 350308 | Colletotrichum | lupini | A1 | United Kingdom | Lupinus sp. | AJ300561 | KM251951 | KM252079 | KM252203 | |
| CBS 198.35 [18] | Colletotrichum | kinghornii | A7 | United Kingdom | Phormium sp. | JQ948454 | JQ950105 | KM252083 | JQ948785 | |
| PD93-1748, CBS 126527 [18] | Colletotrichum | godetiae | A4 | United Kingdom | Prunus avium | JQ948408 | JQ950059 | KM252101 | JQ948739 | |
| Isolates from different host worldwide and used as references for genetics groups / species | ||||||||||
| PT250, CBS 129953 [18] | Colletotrichum | rhombiforme | A6 | Portugal | Olea europaea | JQ948457 | JQ950108 | KM251971 | JQ948788* | |
| PT135, CBS 129945 [18] | Colletotrichum | nymphaeae | A2 | Portugal | Olea europaea | JQ948201 | JQ949852 | KM251972 | JQ948531 | |
| PD85-694, CBS 126519 [18] | Colletotrichum | chrysanthemi | A2 | Netherlands | Chrysanthemum sp. | JQ948272 | JQ949923 | KM251973 | JQ948602 | |
| PD89-582, CBS 126524 [18] | Colletotrichum | simmondsii | A2 | Netherlands | Cyclamen sp. | JQ948281 | JQ949932 | KM251974 | JQ948611* | |
| PT227, CBS 129952 [18] | Colletotrichum | acutatum | A5 | Portugal | Olea europaea | JQ948364 | JQ950015 | KM251975 | JQ948695* | |
| Tom-21, CBS 129954 [18] | Colletotrichum | tamarilloi | A8 | Colombia | Cyphomandra betacea | JQ948188 | JQ949839 | KM251976 | JQ948518 | |
| Tom-12, CBS 129955 [18] | Colletotrichum | tamarilloi | A8 | Colombia | Cyphomandra betacea | JQ948189 | JQ949840 | KM251977 | JQ948519 | |
| CBS 193.32 [18] | Colletotrichum | godetiae | A4 | Greece | Olea europaea | JQ948415 | JQ950066 | KM251978 | JQ948746* | |
| PT30 | Colletotrichum | lupini | A1 | Portugal | Lupinus albus | AJ300561 | AJ292250 | KM251979 | KM252117* | |
| CR46, CBS 129947 [18] | Colletotrichum | fioriniae | A3 | Portugal | Vitis vinifera | JQ948343 | JQ949994 | KM251980 | JQ948673* | |
| 9178 | Colletotrichum | salicis | A7 | Norway | Vaccinium corymbosum | KM246583 | KM251937 | KM252051 | KM252187* | |
| MP1, CBS 129972 [18] | Colletotrichum | salicis | A7 | USA | Acer platanoides | JQ948466 | JQ950117 | KM252052 | JQ948797* | |
| PJ8 | Colletotrichum | acutatum | A5 | New Zealand | Pyrus pyrifolia | KM246587 | KM251941 | KM252056 | KM252191* | |
| ATCC MYA-663 | Colletotrichum | fioriniae | A3 | USA | Malus domestica | KM246589 | KM251943 | KM252058 | KM252193* | |
| HY09 | Colletotrichum | lupini | A1 | Canada | Lupinus albus | KJ018635 | KJ018647 | KM252062 | KJ018659* | |
| JL198 | Colletotrichum | godetiae | A4 | Serbia | Olea europaea | AJ749689 | AJ748613 | KM252068 | KM252197* | |
| AR3787, CBS 118191 [18] | Colletotrichum | phormii | A7 | South Africa | Phormium sp. | JQ948453 | JQ950104 | KM252082 | JQ948784* | |
| CBS 607.94 [18] | Colletotrichum | salicis | A7 | Netherlands | Salix sp. | JQ948460 | JQ950111 | KM252085 | JQ948791* | |
| ALM-NRB-30K | Colletotrichum | godetiae | A4 | Israel | Prunus dulcis | DQ003129 | KM251960 | KM252094 | KM252212* | |
| CBS 101611 [18] | Colletotrichum | sp. 1 | - | Costa Rica | Fern | JQ948196 | JQ949847 | KM252095 | JQ948526* | |
| BBA 70884, CBS 109225 [18] | Colletotrichum | lupini | A1 | Ukraine | Lupinus albus | JQ948155 | JQ949806 | KM252096 | JQ948485* | |
| STE-U 164, CBS 112980 [18] | Colletotrichum | acutatum | A5 | South Africa | Pinus radiata | JQ948356 | JQ950007 | KM252097 | JQ948687* | |
| STE-U 5303, CBS 112989 [18] | Colletotrichum | laticiphilum | A2 | India | Hevea brasiliensis | JQ948289 | JQ949940 | KM252098 | JQ948619 | |
| CBS 122122 [18] | Colletotrichum | simmondsii | A2 | Australia | Carica papaya | JQ948276 | JQ949927 | KM252099 | JQ948606* | |
| CBS 211.78 [18] | Colletotrichum | costaricense | - | Costa Rica | Coffea sp. | JQ948181 | JQ949832 | KM252102 | JQ948511 | |
| DPI 11711, CBS 292.67 [18] | Colletotrichum | brisbanense | A2 | Australia | Capsicum annuum | JQ948291 | JQ949942 | KM252103 | JQ948621 | |
| DPI 13483, CBS 294.67 [18] | Colletotrichum | simmondsii | A2 | Australia | Carica papaya | JQ948277 | JQ949928 | KM252104 | JQ948607* | |
| ATCC 38896, CBS 526.77 [18] | Colletotrichum | nymphaeae | A2 | Netherlands | Nymphaeae alba | JQ948199 | JQ949850 | KM252105 | JQ948529 | |
| CBS 797.72 | Colletotrichum | fioriniae | A3 | New Zealand | Pinus radiata | KM246598 | KM251961 | KM252106 | KM252213* | |
| OCO-ARC-4 | Colletotrichum | sp. 2 | - | USA | Citrus x sinensis | EU647305 | KM251962 | KM252107 | EU647318* | |
| STF-FTP-10 | Colletotrichum | sp. 2 | - | USA | Citrus x sinensis | EU647306 | KM251963 | KM252108 | EU647319 | |
| Coll-25 | Colletotrichum | scovillei | A2 | Taiwan | Capsicum annum | KJ018637 | KJ018649 | KM252109 | KJ018661 | |
| Coll-154 | Colletotrichum | scovillei | A2 | Taiwan | Capsicum annum | DQ410028 | KM251964 | KM252110 | KM252214 | |
| Isolates as out-group | ||||||||||
| CSL 311 | Colletotrichum | fruticola | OG | USA | Fragaria x ananassa | KM246512 | KM251865 | KM251965 | KM252111* | |
| CSL 386 | Colletotrichum | fruticola | OG | USA | Fragaria x ananassa | KM246513 | KM251866 | KM251966 | KM252112* | |
| CSL 780 | Colletotrichum | aenigma | OG | UK | Fragaria x ananassa | KM246517 | KM251871 | KM251984 | KM252121* | |
| CSL 869 | Colletotrichum | aenigma | OG | UK | Fragaria x ananassa | KM246540 | KM251895 | KM252008 | KM252145* | |
| CSL 593 | Colletotrichum | spinaciae | OG | UK | Spinacia oleracea | KM246596 | KM251958 | KM252092 | KM252210 | |
| CSL 739 | Colletotrichum | spinaciae | OG | UK | Spinacia oleracea | KM246597 | KM251959 | KM252093 | KM252211 | |
| M1.001 [27] | Colletotrichum | graminicola | OG | USA | Zea mais | genome: ACOD0100000000 | ||||
| IMI 349063 [27] | Colletotrichum | higginsianum | OG | Trinidad and Tobago | Brassica chinensis | genome: CACQ0200000000 | ||||
Abbreviation
CBS: Culture collection of the Centraalbureau voor Schimmelcultures, Fungal Biodiversity Centre, Utrecht, The Netherlands IMI: Culture collection of CABI Europe UK Centre, Egham, UK CSL: Culture collection of The Food and Eviroment Research Agency, DEFRA, York, UK OG: out-group* strains used for pathogenicity tests
Cultures were maintained at 25°C on potato dextrose agar medium (PDA, Difco Laboratories, USA) for up to ten days under a 12 h light/ 12 h dark cycle. Long–term storage at 4°C involved cutting mycelial plugs from the edge of actively growing cultures on PDA and suspending them in sterile water.
Characterization of genetic variation
Genomic DNA was extracted according to the Chelex 100 protocol [29], with some modifications [30]. DNA was quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, DE, USA).
Various target regions were used to characterise genetic diversity amongst the fungal isolates including: ITS region, partial sequence of the beta-tubulin 2 gene (TUB) (exons 3 through 6, including introns 2 through 4), partial sequence of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene, and partial sequence of the mating type gene (MAT1-2) (the intron included in the conserved HMGbox region). Target regions were amplified using PCR reaction mixes (20 μl) that contained 1 μl of DNA, 1 μl each of primer (20 μM), 7 μl of H20 and 10 μl of ReadyMix RedTaq (Sigma).
PCR amplification of the target regions for sequencing was carried out as described below using previously published primers under conditions standardised for routine work. For ITS, primers ITS1Ext and ITS4Ext [31] were used. The amplification program consisted of 2 min of initial denaturation (95°C), 30 cycles of amplification (1 min at 94°C, 1 min at 55°C, and 1 min at 72°C) and a final extension at 72°C for 5 min. For TUB, primers TB5 and TB6 [31] were used. The amplification program consisted of 2 min initial denaturation (95°C), 30 cycles of amplification (1 min at 94°C, 1 min at 65°C and 1 min at 72°C) and a final extension at 72°C for 5 min. For GAPDH, primers GDF1 and GDR1 [32] were used. The amplification program consisted of 2 min initial denaturation at 95°C, 35 cycles of amplification (1 min at 94°C, 1 min at 60°C and 30 sec at 72°C) and a final extension at 72°C for 3 min. For MAT1-2, primers HMGacuF2 and HMGacuR [21] for C. acutatum s.l. and primers HMGgloeF1 and HMGgloeR1 for C. gloeosporioides s. l. [33] were used. The amplification program consisted of 5 min initial denaturation at 95°C, 40 cycles of amplification (1 min at 95°C, 1 min between 48°C and 55°C and 30s at 72°C) and a final extension of 20 min at 72°C. PCR products were separated using gel electrophoresis and purified using the QIAquick PCR purification kit (Qiagen, USA).
Sequencing of PCR products was carried out at the University of Warwick Genomics Centre, using an ABI Prism 7900HT or ABI3100 sequence detection system (Applied Biosystems, UK). PCR products were cleaned up and then quantified with reference to a ladder (Bioline EasyLadder I) containing DNA fragments of known concentration. One to five microliters of each sample (depending on DNA concentration) were used in sequencing reactions with the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, UK). ABI trace files were analyzed and consensus sequences were generated using Geneious 7.1.6 [34]. All the sequences were aligned using MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/) and were manually edited to optimise the alignment, as required. Multiple alignments were end trimmed in order to have comparable nucleotides.
Multiple sequence alignments were exported to MEGA5 [35] where best-fit substitution models were calculated for each separate sequence dataset. In order to evaluate whether the four sequenced loci were congruent and suitable for concatenation, tree topologies of 50% Neighbour-Joining bootstrap and maximum parsimony analysis (100,000 replicates) were separately performed for each gene and visually compared [36]. The multilocus concatenated alignment (ITS, TUB2, MAT1-2 and GAPDH) was performed with Geneious 7.1.6 [34]. A Markov Chain Monte Carlo (MCMC) algorithm was used to generate phylogenetic trees with Bayesian probabilities using MrBayes 3.2.1 [37] for combined sequence datasets. Models of nucleotide substitution for each gene determined by MEGA5 were included for each locus. The analysis in MrBayes ran for 5000000 of generations to reach a P value lower than 0.01 with two parallel searches using three heated and one cold Markov chain sampled every 100 generations; 25% of generations were discarded as burn-in. Further phylogenetic analysis was performed by the neighbour-joining method with 1,000 bootstrap replicates under Kimura’s two-parameter correction using Geneious 7.1.6 [34] and the results are presented in Figs 1 and 2.
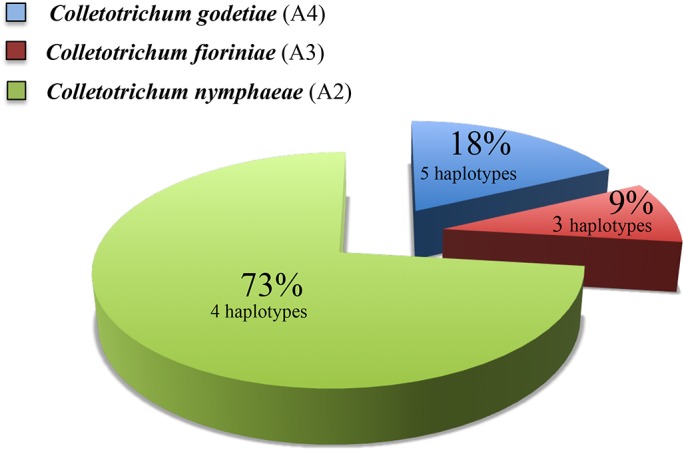
Fig 2. Percentage occurrence of Colletotrichum acutatum sensu lato species and relative numbers of haplotypes identified among 67 strains isolated from strawberry in UK.
Comparison of fungal growth in culture
The 67 fungal isolates collected from strawberry in the UK were compared with a subset of other isolates (chosen based on genetic, host and geographic diversity) including 49 isolates of C. acutatum s.l. and four isolates of C. gloeosporioides s.l. for in vitro growth studies on PDA (Potato Dextrose Agar, BD Difco). For experiments, a 7 mm diameter mycelial plug excised from the edge of an actively growing PDA culture was placed at the centre of a fresh PDA plate. In the growth experiment, two perpendicular colony diameters were measured daily and colony radius was calculated from cultures incubated at four different temperatures (15°C, 20°C, 25°C and 30°C) in darkness. Data corresponding to the linear growth phase were subjected to analysis of variance of regression in order to create growth curves for each isolate at each temperature. In both tests three plates were used as replicates. Statistical analysis was performed by SIGMAPLOT 10 program (Sigmaplot Software, USA). Colony characters were recorded after 15 days of incubation at 25°C under 12 h light/ 12 h dark cycle.
Pathogenicity tests
Representative isolates (highlighted with asterisks in Table 1) of each C. acutatum s.l. group isolated from strawberry in UK, together with reference isolates from other hosts, were used for pathogenicity tests on the generally susceptible strawberry cultivar Elsanta [38]. A conidial suspension was prepared for each isolates by flooding 10-day-old PDA culture plates with sterile deionised water. Spore concentration was adjusted to 105 spores ml-1 and 106 spores ml-1 for fruit and crown inoculation, respectively [7,38]. Unripe fruits (white fruit beginning to turn pink, as shown in Fig 3A) [39] were inoculated with a 5μl drop of conidial suspension. Before inoculation, fruit surfaces were disinfected for 5 min using NaClO (1% active chlorine) in 50% EtOH, washed three times in sterilized water, blotted dry and placed in a tray with moist sand on the bottom to prevent movement of the fruits during further procedures. After inoculation, fruits were incubated at 25°C under 12h light/ 12h in dark cycle.
Fig 3. Strawberry fruits and plants used for pathogenicity tests (A and C) and symptoms (B and D).
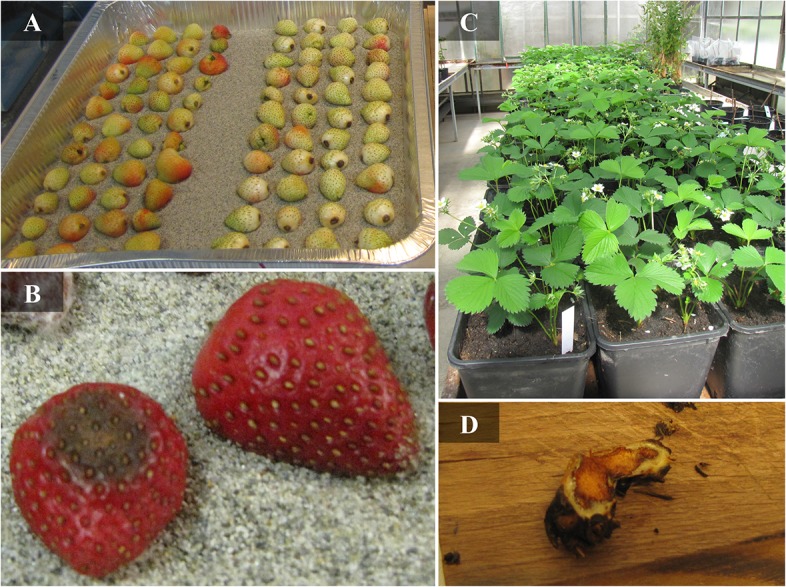
(A) Unripe fruits (phenological stage turning white-pink) used for artificial inoculations of Colletotrichum spp. (B) Strawberry fruits 7 days after inoculation with Colletotrichum sp. spores suspension showing typical black spot symptoms (bottom left) and with sterile water used as control (top right) (C) Three-month-old strawberry plants used to pathogenicity assays (D) Strawberry plant crown sectioned showing presence of red-brownish lesions characteristic of anthracnose caused by Colletotrichum spp.
Disease symptoms were evaluated 7 days after inoculation (d.a.i.) (Fig 3B) by recording the incidence of disease (% of infected fruits), and the aggressiveness of lesion development using the following severity scale: 0, no visible lesions; 1, lesions on less than 33% of fruit surface; 2, lesions covering 33–66% of fruit surface; and 3, lesions covering more than 66% of fruit surface. Three fruits inoculated with sterile distilled water (SDW) as well as fresh fruits served as non-inoculated controls. Four independent replicates were tested for each fungal isolate, consisting of three inoculated fruits for each replicate. At the end of the experiment, Colletotrichum isolates were re-isolated from infected fruits and cultured on PDA to confirm colony characteristics.
The capability of the isolates to produce crown rot symptoms was evaluated by injecting the crowns of three-months-old strawberry plants (Fig 3C) with 0.2 mL conidial suspension using a syringe [4,7]. Plants were placed in glasshouse at 23°C with 16h light / 8h darkness. After 24 days (d.a.i.), plants were evaluated for the presence of crown tissues with red-brownish discoloration, wilting and collapse of the plant, typical symptoms of Colletotrichum crown rot, according to the following severity scale: 0, no lesions; 1, crown tissues discoloration but no wilting or collapse; 2, wilting or collapse of part of the plant; and 3, plant death. Crowns of all plants were sectioned and examined for the presence of red-brownish lesions (Fig 3D). Crown infection was confirmed by re-isolation of the pathogen. Three plant crowns injected with SDW as well as untouched plants served as negative controls for each replicates. The experiment was independently replicated three times, with six plants for each replicate.
Values of disease severity were used to calculate a Disease Index (DI, average severity) according to the following formula: Σvn/N, where v represents the numeric value of the class, n is the number of plants or fruits assigned to the class, N is the total number of the plants or fruits assessed. Data for pathogenicity tests on both fruits and plants were subjected to analysis of variance ANOVA and means compared using Tukey’s multiple range test by Systat11 (Systat Software, USA).
Results
Characterization of genetic variation, and species identification
Phylogenetic trees were constructed using combined ITS, TUB2, GADPH and MAT1-2 sequence data set consisting of 148 Colletotrichum isolates (Table 1). As shown in Fig 1, most of the C. acutatum s.l. isolates (49/67) were identified as belonging to C. nymphaeae (= A2 genetic group), based on clustering with high bootstrap value with the reference isolates CBS 797.72, PT135, IMI345028 and other genetically similar isolates (identical sites = 1422/1438 or 98.9%; pairwise identity = 99.9%). A smaller proportion of isolates in the diversity collection (12/67) were identified as belonging to C. godetiae (= A4 genetic group) based on genetic clustering with reference isolates ALMNRB-30K, CBS 193.32 and JL198 (identical sites = 1411/1438 or 94.6%; pairwise identity = 99.4%). And finally, six isolates were identified as belonging to C. fioriniae (= A3 genetic group) based on clustering with the reference isolate ATCC 56813 (identical sites = 1.436 /1443 or 99.5%; pairwise identity = 99.9%).
Molecular characterisation of 67 Colletotrichum isolates collected from strawberry in the UK along with the reference isolates representing the host and geographic diversity (Figs 1 and 2) suggests that there have been multiple introductions of the anthracnose pathogen belonging to different Colletotrichum species into the country. Three different species C. nymphaeae, C. godetiae and C. fioriniae were identified based on sequence from four loci [6,17,18]. Incidence of these species is shown in Fig 2, where C. nymphaeae corresponds to 73%, followed by C. godetiae (18%) and C. fioriniae (9%). GAPDH is the locus that shows the highest variability across the nucleotide dataset, with 24.1% identical sites for the entire set of data (out-group included) and 59.3% within C. acutatum s.l. The MAT1-2 gene also shows a high variability with 34.4% identical sites of which 78.6% in C. acutatum s.l. TUB and ITS loci show lower percentage of variable sites. In detail, TUB has 58.1% of identical sites in the final alignment and 80.7% only considering C. acutatum s.l. While ITS has 77.8% and 92.4% of conserved nucleotides, respectively with and without out-groups. Based on the nucleotide variability referred to above, four haplotypes of C. nymphaeae, three haplotypes of C. fioriniae, and five haplotypes of C. godetiae were identified further highlighting the multiple introductions of the pathogens belonging to these species into the UK.
Fungal growth in plate culture
Radial growth data of C. acutatum s.l. and C. gloeosporioides s.l. isolates were subjected to analysis of variance of regression in order to obtain growth curves that were all statistically significant (R2≥0.9447 and P<0.0001), with the only exception of one isolate showing a R2 = 0.770 (C. nymphaeae CSL224 at 30°C). The slope for each isolate (three replicates for each isolate) belonging to the same species were averaged, in order to detect the hypothetical optimal growth temperature, and results are shown in Table 2. Almost all species, particularly those containing isolates from strawberry in the UK namely C. nymphaeae, C. fioriniae, and C. godetiae had highest growth rates at 25°C that was considered as optimum temperature. It is pertinent to mention that higher levels of strawberry anthracnose incidence in the UK have been reported in the southwest and southeast regions, where relatively high temperatures are most often reached [20]. However, C. phormii, C. kinghormii and C. rhombiforme showed the highest growth rate at the temperature of 20°C and they were not able to grow at 30°C. Interestingly, these three species are evolutionarily closely related, suggesting a specific adaptation to different environmental conditions compared to other members of the same complex. With respect to C. gloeosporioides s.l. isolates (C. aenigma CSL780 and CSL 869; C. fruticola CSL 311 and CSL386), used as out-groups, all the four isolates showed the highest growth rate at all the tested temperatures when compared with all the other isolates.
Table 2. Radial growth rate (mm h-1) of each Colletotrichum species at different temperatures.
| Species | 15°C* | 20°C* | 25°C* | 30°C* | |
|---|---|---|---|---|---|
| out-group | C. aenigma | 0.112 ± 0.001 | 0.199 ± 0.002 | 0.261 ± 0.008 | 0.124 ± 0.011 |
| C. fruticola | 0.118 ± 0.004 | 0.209 ± 0.005 | 0.238 ± 0.019 | 0.150 ± 0.008 | |
| Colletotrichum acutatum species complex | C. rhombiforme | 0.091 ± 0.001 | 0.135 ± 0.001 | 0.111 ± 0.002 | 0.000 ± 0.000 |
| C. kinghornii | 0.073 ± 0.001 | 0.108 ± 0.001 | 0.077 ± 0.002 | 0.000 ± 0.000 | |
| C. phormii | 0.106 ± 0.001 | 0.166 ± 0.001 | 0.139 ± 0.002 | 0.000 ± 0.000 | |
| C. salicis | 0.094 ± 0.001 | 0.147 ± 0.002 | 0.179 ± 0.004 | 0.035 ± 0.005 | |
| C. godetiae | 0.094 ± 0.002 | 0.142 ± 0.003 | 0.163 ± 0.005 | 0.004 ± 0.000 | |
| C. acutatum | 0.054 ± 0.004 | 0.087 ± 0.006 | 0.148 ± 0.004 | 0.058 ± 0.005 | |
| C. fioriniae | 0.081 ± 0.003 | 0.136 ± 0.005 | 0.185 ± 0.004 | 0.083 ± 0.006 | |
| Colletotrichum sp. 2 | 0.085 ± 0.002 | 0.140 ± 0.001 | 0.178 ± 0.002 | 0.075 ± 0.002 | |
| C. lupini | 0.086 ± 0.001 | 0.130 ± 0.003 | 0.152 ± 0.009 | 0.058 ± 0.002 | |
| Colletotrichum sp. 1 | 0.083 ± 0.001 | 0.132 ± 0.001 | 0.138 ± 0.001 | 0.043 ± 0.001 | |
| C. tamarilloi | 0.069 ± 0.001 | 0.123 ± 0.002 | 0.148 ± 0.003 | 0.007 ± 0.000 | |
| C. simmondsii | 0.040 ± 0.003 | 0.092 ± 0.008 | 0.112 ± 0.014 | 0.089 ± 0.010 | |
| C. laticiphilum | 0.058 ± 0.002 | 0.113 ± 0.001 | 0.161 ± 0.001 | 0.121 ± 0.002 | |
| C. nymphaeae | 0.077 ± 0.001 | 0.135 ± 0.002 | 0.159 ± 0.004 | 0.063 ± 0.005 | |
| C. chrysanthemi | 0.050 ± 0.001 | 0.083 ± 0.001 | 0.111 ± 0.001 | 0.087 ± 0.002 | |
| C. scovillei | 0.036 ± 0.001 | 0.105 ± 0.001 | 0.115 ± 0.001 | 0.062 ± 0.002 |
* Values represent the average + SD of slopes (growth rates expressed as mm h-1) of all isolates belonging to the same species, three replicates for each isolate. The optimal temperature for each species is indicated in bold.
C. nymphaeae isolates developed white cottony aerial mycelium, light brownish conidial masses with peculiar colony colour from dark grey to dark brown. Twelve isolates belonging to C. godetiae were characterized by white aerial mycelium, and yellow pigmentation to white colour on the reverse side of the culture. C. fioriniae isolates were dark red on the reverse side of the cultures with orange conidial masses in large drops on the colony surface, and conidiomata formed directly on the hyphae. However, these characters are often difficult to describe reliably, and can change following sub-culturing or based on the length and type of storage. Thus, there is a need for further development of molecular methods for reliable and rapid diagnosis and monitoring of the pathogen populations belonging to different species associated with strawberry production in a specific geographic location.
Characterisation of variation in pathogenicity
Thirty-four C. acutatum s.l. isolates were chosen for pathogenicity tests on fruits and plants, including six representative isolates from each of the three species described above related to strawberry production in the UK (highlighted with * in Table 1 and in bold in Fig 1), and one or more isolates representative of all the major species of the C. acutatum complex. Four C. gloeosporioides s.l. isolates that were isolated from strawberry infected tissues from UK (CSL 780 and CSL 869, C. aenigma) and USA (CSL 311 and CSL 386, C. fruticola) were included in the experiments as an out-group.
C. acutatum s.l. isolates varied in aggressiveness on both host tissues. In the fruit assays, among the three species identified from the strawberry production systems in the UK, C. nymphaeae and C. fioriniae were more aggressive compared to C. godetiae. This was particularly noticeable for isolates originating from strawberry as reflected by the fruit disease index range for C. nymphaeae (2.08–3.00), C. fioriniae (1.92–2.75) and C. godetiae (0.75–2.08). Interestingly, with isolates originating from other hosts, C. nymphaeae isolates were less aggressive (0.67–1.67), and one or more isolates belonging to C. fioriniae (2.00–2.17) as well as C. godetiae (2.17) showed fruit disease index in the range of the strawberry isolates. Among the other species tested within the C. acutatum complex, C. acutatum s.s., C. simmondsii and Colletotrichum sp.2 included one or more isolates originating from non-strawberry hosts that showed medium level of aggressiveness with fruit disease index ranging from 1.17 to 2.08. Whereas, C. lupini (0.08–075), C. phormii (0.58), C. salicis (0.17–0.67), and C. rhombiforme (0.67) along with Colletotrichum sp.1 (0.33) isolates originating from various hosts other than strawberry were much less aggressive as reflected by the fruit disease index. The C. gloeosporioides s.l. isolates tested showed a fruit disease index ranging from 1.50 to 2.50 (Table 3).
Table 3. Variability in aggressiveness of Colletotrichum species isolates on strawberry fruits and plants.
| Isolate | Species | Isolation source | Origin | Fruit Disease index * , + | Plant Disease Index * , # | |||
|---|---|---|---|---|---|---|---|---|
| Colletotrichum acutatum species complex | CSL 256 | C. nymphaeae | Fragraria | UK | 2.50 | abcd | 0.50 | bc |
| CSL 899 | C. nymphaeae | Fragraria | UK | 3.00 | a | 0.83 | abc | |
| CSL 915 | C. nymphaeae | Fragraria | UK | 2.08 | abcdef | 0.61 | bc | |
| ATCC 38896 | C. nymphaeae | Nymphaeae | Netherlands | 0.67 | defg | 0.28 | bc | |
| CSL 455 | C. nymphaeae | Photinia | UK | 1.08 | bcdefg | 0.56 | bc | |
| PT135 | C. nymphaeae | Olea | Portugal | 1.67 | abcdefg | 0.89 | abc | |
| CSL 916 | C. godetiae | Fragraria | UK | 1.92 | abcdefg | 0.39 | bc | |
| CSL 918 | C. godetiae | Fragraria | UK | 0.75 | cdefg | 0.39 | bc | |
| CSL 919 | C. godetiae | Fragraria | UK | 2.08 | abcdef | 0.67 | bc | |
| ALM-NRB-30K | C. godetiae | Prunus | Israel | 0.25 | fg | 0.11 | c | |
| CBS 193.32 | C. godetiae | Olea | Greece | 0.75 | cdefg | 0.28 | bc | |
| JL198 | C. godetiae | Olea | Serbia | 2.17 | abcde | 0.39 | bc | |
| CSL 1259 | C. fiorinae | Fragraria | UK | 2.75 | ab | 0.72 | bc | |
| CSL 1262 | C. fiorinae | Fragraria | UK | 1.92 | abcdefg | 1.00 | ab | |
| CSL 2546 | C. fiorinae | Fragraria | UK | 2.67 | abc | 0.72 | bc | |
| CBS 797.72 | C. fiorinae | Pinus | New Zealand | 1.08 | bcdefg | 0.39 | bc | |
| ATCC MYA-663 | C. fiorinae | Malus | USA | 2.00 | abcdef | 0.83 | abc | |
| CR46 | C. fiorinae | Vitis | Portugal | 2.17 | abcde | 0.33 | bc | |
| PJ8 | C. acutatum | Pyrus | New Zealand | 2.08 | abcdef | 0.72 | bc | |
| PT227 | C. acutatum | Olea | Portugal | 1.42 | abcdefg | 0.78 | abc | |
| STE-U-164 | C. acutatum | Pinus | South Africa | 0.83 | cdefg | 0.28 | bc | |
| CBS 122122 | C. simmondsii | Carica | Australia | 0.25 | efg | 0.22 | bc | |
| CBS 294.67 | C. simmondsii | Carica | Australia | 1.17 | abcdefg | 0.61 | bc | |
| PD89-582 | C. simmondsii | Cyclamen | Netherland | 1.83 | abcdefg | 0.44 | bc | |
| BBA 70884 | C. lupini | Lupinus | Ukraine | 0.58 | efg | 0.33 | bc | |
| HY09 | C. lupini | Lupinus | Canada | 0.08 | g | 0.17 | bc | |
| PT30 | C. lupini | Lupinus | Portugal | 0.75 | cdefg | 0.56 | bc | |
| 9178 | C. salicis | Vaccinium | Norway | 0.50 | efg | 0.28 | bc | |
| CBS 607.94 | C. salicis | Salix | Netherlands | 0.67 | defg | 0.17 | bc | |
| MP1 | C. salicis | Acer | USA | 0.17 | fg | 0.22 | bc | |
| CBS 101611 | Colletotrichum sp. 1 | Fern | Costa Rica | 0.33 | efg | 0.06 | c | |
| OCO-ARC-4 | Colletotrichum sp. 2 | Citrus | USA | 1.42 | abcdefg | 0.11 | c | |
| AR3787 | C. phormii | Phormium | South Africa | 0.58 | efg | 0.22 | bc | |
| PT250 | C. rhombiforme | Olea | Portugal | 0.67 | defg | 0.33 | bc | |
| out-group | CSL 780 | C. aenigma | Fragraria | UK | 2.50 | abcd | 0.50 | bc |
| CSL 869 | C. aenigma | Fragraria | UK | 1.92 | abcdefg | 0.72 | bc | |
| CSL 311 | C. fruticola | Fragraria | USA | 2.50 | abcd | 1.56 | a | |
| CSL 386 | C. fruticola | Fragraria | USA | 1.50 | abcdefg | 0.22 | bc | |
Disease Index data related to aggressiveness on strawberry fruits and crowns of representative Colletotrichum isolates.
*: Different letters within the same column correspond to significantly different values (ANOVA; P < 0.05). The values are the averages ± SD of four independent replicates, three fruits for each replicate and of three independent replicates, six plants for each replicate. Disease Index was calculated according to the following formula: Σvn/N, where v represents the numeric value of the class, n is the number of fruits or plants assigned to the class, N is the total number of the plants assessed.
+: 0, no visible lesions; 1, lesions on less than 33% of fruit surface; 2, lesions covering 33–66% of fruit surface; and 3, lesions covering more than 66% of fruit surface.
#: 0, no lesions; 1, crown tissues discoloration but no wilting or collapse; 2, wilting or collapse of part of the plant; and 3, plant death.
In the in vitro assays, anthracnose fruit rot symptoms were observed (e.g. Fig 3B) for various isolates tested with different levels of aggressiveness, as shown by the disease index ranging from 0.08 to 3.0 (Table 3). The variation in aggressiveness among different isolates was clearly reflected by the differences in incidence which ranged from 8.33 to 100% with only 4 out of 38 isolates showing 91.7 to 100% as well as the lesion type which ranged from 0.1 to 3.0 (S1 Table). When lesion morphology was evaluated, different kinds of lesions could be distinguished on fruits, ranging from brown ones containing orange drops of conidia to those entirely covered with aerial mycelium, with different lesion size. C. nymphaeae CSL899 was the most aggressive on strawberry fruits with the highest disease index (3.0, corresponding to symptoms covering more than 66% of fruit surface).
In the plant assays, varying degrees of crown rot symptoms were recorded 24 d.a.i, as reflected by the disease index range shown in Table 3. Symptom severity was generally low, with no isolate scoring higher than 2 (wilting and collapse of plant). Among the three species identified from UK strawberry production systems, C. fioriniae isolates originating from strawberry showed a higher range of disease index (0.72–1.00) compared to C. nymphaeae (0.5–0.83) and C. godetiae (0.39–0.67). The C. gloeosporioides s.l. isolate CSL 311 (C. fruticola from strawberry in USA) showed the highest disease index (1.6), this isolate was also amongst the most aggressive on fruit (Table 3). Colletotrichum isolates were recovered from all crowns showing symptoms.
Discussion
The UK strawberry industry has expanded rapidly in recent years, and this appears to correlate with increasing losses attributed to anthracnose caused by Colletotrichum spp. [6]. This study provides the first molecular characterization of C. acutatum sensu lato diversity related to strawberry production in the UK, combined with pathogenic characterization. A collection of 148 isolates representative of UK and global diversity of C. acutatum s.l. populations has been assembled. The isolates were chosen based on host association, geographic distribution, phylogenetic relationships and biological diversity.
On the basis of four sequence loci (ITS, TUB, GAPDH, and MAT1-2), the C. acutatum sensu lato isolates were assigned to three newly designated species C. nymphaeae, C. godetiae and C. fioriniae following a recent taxonomic re-assessment [18]. According to available literature, C. nymphaeae is the most common and C. godetiae is also often reported in European and American strawberry fields [6]. These two species were also the most representative in our dataset of isolates related to strawberry in the UK. C. fioriniae has a worldwide distribution and is common on strawberry but only a few isolates were identified in our collection, and this group was not commonly present in the fields in the UK. C. simmondsii, C. acutatum sensu stricto, C. salicis and C. miyabeana are common on strawberry in Oceania and have only been found sporadically in Europe. Isolates belonging to these species have not been detected on strawberry in the UK. The variability observed within the UK C. acutatum sensu lato species fits in part with previous reports of C. acutatum on strawberry within specific geographic regions. For example, in France, Israel, Bulgaria and Spain, the majority of strawberry anthracnose pathogen isolates clustered in the same species C. nymphaeae, and almost no intra-specific diversity was observed within each country [2–5]. A different situation has been observed on Belgian isolates, where the population represented: 33% isolates belonging to C. nymphaeae, 5% C. fioriniae, 50% C. godetiae, 3% C. acutatum s.s. and 6% C. salicis. A possible explanation to C. acutatum s.l. status in the UK might be recent introduction (late 70s) from a limited number of sources. The reason for the differences in the occurrence of various Colletotrichum species associated with strawberry production in different geographic locations still remains unclear, but the source of importation of the planting material and local trade have been heavily implicated [4,7].
The pathogenicity assays used in this work are based on a study in Belgium [7] in view of the similar molecular diversity of the anthracnose pathogen populations associated with strawberry production. These assays with the isolates representing the molecular diversity not only revealed variability in aggressiveness in different species described within C. acutatum s.l., but also complex patterns both between and within the species. For example, based on isolates originating from strawberry, C. fioriniae and C. nymphaeae appear equally aggressive on fruits with C. nymphaeae isolates indicating a degree of host-preference. Both C. fioriniae and C. godetiae included isolates originating from other hosts that showed comparable levels of aggressiveness to isolates from strawberry. Similar situation was observed with at least some non-strawberry isolates belonging to species such as C. acutatum s.s. and C. simmondsii. Furthermore, at least one C. godetiae isolate from strawberry was much less aggressive compared to others. These patterns suggest that some Colletotrichum species such as C. fioriniae and C. godetiae include populations that are capable of infecting a wider range of hosts, also influenced by environmental conditions. Further studies using a wider set of isolates of these three species and appropriate pathological and biological assays are required to gain additional insights into the evolution of pathogenicity in relation to field symptoms as well as any differential responses to host varieties and fungicides locally used in the UK strawberry production systems.
The study has highlighted the genetic and pathogenic heterogeneity of the introduced anthracnose pathogen populations belonging to three different Colletotrichum species emphasising the need for effective phytosanitary procedures linked to pathogen monitoring and characterisation to generally limit the entry of non-native pathogens. This also underlines the requirement of reliable and rapid diagnostic tools for further research and application in strawberry anthracnose management. The recent release of a whole genome sequence of C. fioriniae isolated from strawberry [28] along with the newly characterised isolates, based on multi-locus sequence and aggressiveness information reported here, represents a useful platform for further research into the genetic basis of C. acutatum s.l.—strawberry interactions.
Supporting Information
a 0, no visible lesions; 1, lesions on less than 33% of fruit surface; 2, lesions covering 33–66% of fruit surface; and 3, lesions covering more than 66% of fruit surface. b no lesions; 1, crown tissues discoloration but no wilting or collapse; 2, wilting or collapse of part of the plant; and 3, plant death.
(XLSX)
Acknowledgments
The authors would like to dedicate this work to Maurizio Forti (University of Pisa), who passed away in December 2013 and to Dez Barbara (University of Warwick) who passed away in July 2012. The authors would like to thank Fera and the University of Warwick for funding this research and providing the strains set. They are especially thankful to: Ulrike Damm (CBS-KNAW Fungal Biodiversity Centre–The Netherlands), Paul Cannon and Alan Buddie (CABI—UK), Gunn Mari Strømeng (Norwegian University of Life Sciences—Norway), Katherine LoBuglio (Harvard University Herbaria, USA), Peter R. Johnston (Manaaki Whenua Landcare Research–New Zealand), James Cunnington (Institute for Horticultural Development—Australia), Amy Rossman (USDA-ARS–USA), Stanley Freeman (ARO Volcani Center—Israel), Daniel Buchvaldt Amby (University of Copenhagen–Denmark), Natalia Peres (University of Florida–USA) and Sheu Zong-ming (AVRDC–The World Vegetable Center–Taiwan) for kindly providing reference isolates.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors would like to thank University of Warwick for funding this research. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Beech MG, Simpson DW. Strawberry production in the United Kingdom. ISHS Acta Horticulturae. 1989;265: 693–696. [Google Scholar]
- 2. Denoyes-Rothan B, Guerin G, Delye C, Smith B, Minz D, Maymon M, et al. Genetic diversity and pathogenic variability among isolates of Colletotrichum species from strawberry. Phytopathology. 2003. ; 93(2): 219–28. 10.1094/PHYTO.2003.93.2.219 [DOI] [PubMed] [Google Scholar]
- 3. Freeman S, Katan T. Identification of Colletotrichum Species Responsible for Anthracnose and Root Necrosis of Strawberry in Israel. Phytopathology. 1997;87(5): 516–21. [DOI] [PubMed] [Google Scholar]
- 4. Garrido C, Carbu M, Fernandez-Acero FJ, Budge G, Vallejo I, Colyer A, et al. Isolation and pathogenicity of Colletotrichum spp. causing anthracnose of strawberry in south west Spain. European Journal of Plant Pathology. 2008;120(4): 409–415. [Google Scholar]
- 5. Jelev ZJ, Bobev SG, Minz D, Maymon M, Freeman S. Characterization of Colletotrichum species causing strawberry anthracnose in Bulgaria. Journal of Phytopathology. 2008;156: 668–677. [Google Scholar]
- 6. Sreenivasaprasad S, Talhinhas P. Genotypic and phenotypic diversity in Colletotrichum acutatum, a cosmopolitan pathogen causing anthracnose on a wide range of hosts. Molecular Plant Pathology. 2005;6(4): 361–378. 10.1111/j.1364-3703.2005.00291.x [DOI] [PubMed] [Google Scholar]
- 7. Van Hemelrijck W, Debode J, Heungens K, Maes M, Creemers P. Phenotypic and genetic characterization of Colletotrichum isolates from Belgian strawberry fields. Plant Pathology. 2010;59(5): 53–861. [Google Scholar]
- 8. Weir BS, Johnston PR, Damm U. The Colletotrichum gloeosporioides species complex. Studies in Mycology. 2012;73: 115–180. 10.3114/sim0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Howard CM, Albregts EE. Anthracnose In: Compendium of Strawberry Diseases. American Phytopathological Society: Paul St, ed. Maas JL USA; 1984. pp. 85–87. [Google Scholar]
- 10. Maas JL, Howard CM. Variation of several anthracnose fungi in virulence to strawberry and apple. Plant Disease. 1985;69: 164–166. [Google Scholar]
- 11. Sutton BC. Colletotrichum: Biology, Pathology and Control Bailey JA and Jeger MJ, editors. Wallingford: CAB International, UK; 1992. [Google Scholar]
- 12. Buddie AG, Martinez-Culebras P, Bridge PD, García MD, Querol A, Cannon PF, et al. Molecular characterization of Colletotrichum strains derived from strawberry. Mycological Research. 1999;103: 385–394. [Google Scholar]
- 13. Smith BJ, Black LL. First report of Colletotrichum acutatum on strawberry in the United States. Plant Disease. 1986;70: 1074. [Google Scholar]
- 14. Sreenivasaprasad S, Sharada K, Brown AE, Mills PR. PCR-based detection of Colletotrichum acutatum on strawberry. Plant Pathology. 1996;45(4): 650–655. [Google Scholar]
- 15. Freeman S, Horowitz S, Sharon A. Pathogenic and non pathogenic lifestyles in Colletotrichum acutatum from strawberry and other plants. Phytopathology. 2001;91: 986–992. 10.1094/PHYTO.2001.91.10.986 [DOI] [PubMed] [Google Scholar]
- 16. Peres NA, Timmer LW, Adaskaveg JE, Correll JC. Life styles of Colletotrichum acutatum . Plant Disease. 2005;89: 784–796. [DOI] [PubMed] [Google Scholar]
- 17. Whitelaw-Weckert MA, Curtin SJ, Huang R, Steel CC, Blanchard CL, Roffey PE. Phylogenetic relationships and pathogenicity of Colletotrichum acutatum isolates from grape in subtropical Australia. Plant Pathology. 2007;56(3): 448–463. [Google Scholar]
- 18. Damm U, Cannon P, Woudenberg, Crous PW. The Colletotrichum acutatum species complex. Studies in Mycology. 2012;73(1): 37–113. 10.3114/sim0010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jones D, Baker R. Introductions of non-native plant pathogens into Great Britain, 1970–2004. Plant Pathology. 2007;56: 891–910. [Google Scholar]
- 20. Calleja EJ, Ilbery B, Spence NJ, Mills PR. The effectiveness of phytosanitary controls in preventing the entry of Colletotrichum acutatum in the UK strawberry sector. Plant Pathology. 2012;62(2): 266–278. [Google Scholar]
- 21.Baroncelli R. Colletotrichum acutatum sensu lato: From Diversity Study to Genome Analysis. PhD Dissertation, University of Warwick. 2012. Available: http://wrap.warwick.ac.uk/56428/.
- 22. Baroncelli R, Sreenivasaprasad S, Lane CR, Thon MR, Sukno SA. First report of Colletotrichum acutatum sensu lato (Colletotrichum godetiae) causing anthracnose on grapevine (Vitis vinifera) in the United Kingdom. New Disease Reports. 2014;29: 26. [Google Scholar]
- 23. Baroncelli R, Sreenivasaprasad S, Thon MR, Sukno SA. First report of apple bitter rot caused by Colletotrichum godetiae in the United Kingdom. Plant Disease. 2014;98(7): 1000. [DOI] [PubMed] [Google Scholar]
- 24. Howard CM, Maas JL, Chandler CK, Albregts EE. Anthracnose of strawberry caused by the Colletotrichum complex in Florida. Plant Disease. 1992;76: 976–981. [Google Scholar]
- 25. Ureña-Padilla AR, MacKenzie SJ, Bowen BW, Legard DE. Etiology and population genetics of Colletotrichum spp. causing crown and fruit rot of strawberry. Phytopathology. 2002;92: 1245–1252. 10.1094/PHYTO.2002.92.11.1245 [DOI] [PubMed] [Google Scholar]
- 26. Martínez-Culebras PV, Barrio E, García MD, Querol A. Identification of Colletotrichum species responsible for anthracnose of strawberry based on the internal transcribed spacers of the ribosomal region. FEMS Microbiology Letters. 2000;189(1): 97–101. [DOI] [PubMed] [Google Scholar]
- 27. O'Connell RJ, Thon MR, Hacquard S, Amyotte SG, Kleemann J, Torres MF, et al. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nature Genetics. 2012. ; 44(9): 1060–1065. 10.1038/ng.2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baroncelli R, Sreenivasaprasad S, Sukno SA, Thon MR, Holub E. Draft genome sequence of Colletotrichum acutatum sensu lato (Colletotrichum fioriniae). Genome Announcement 2014;2: e00112–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sepp R, Szabo I, Uda H, Sakamoto H. Rapid techniques for DNA extraction from routinely processed archival tissue for use in PCR. Journal of Clinical Pathology. 1994;47(4): 318–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baroncelli R, Sarrocco S, Zapparata A, Tavarini S, Angelini LG, Vannacci G. Characterization and epidemiology of Colletotrichum acutatum sensu lato (C. chrysanthemi) causing Carthamus tinctorius anthracnose. Plant Pathology. 2015;64(2): 375–384. [Google Scholar]
- 31. Talhinhas P, Sreenivasaprasad S, Neves-Martins J, Oliveira H. Genetic and morphological characterization of Colletotrichum acutatum causing anthracnose of lupins. Phytopathology. 2002;92(9): 986–996. 10.1094/PHYTO.2002.92.9.986 [DOI] [PubMed] [Google Scholar]
- 32. Guerber JC, Liu B, Correll JC, Johnston PR. Characterization of diversity in Colletotrichum acutatum sensu lato by sequence analysis of two gene introns, mtDNA and intron RFLPs, and mating compatibility. Mycologia. 2003;95(5): 872–895. [PubMed] [Google Scholar]
- 33. Du MZ, Schardl CL, Nuckles EM, Vaillancourt LJ. Using mating-type gene sequences for improved phylogenetic resolution of Colletotrichum species complexes. Mycologia. 2005;97(3): 641–658. [DOI] [PubMed] [Google Scholar]
- 34. Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12): 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution. 2011;28(10): 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mason-Gamer RJ, Kellogg EA. Testing for phylogenetic conflict among molecular data sets in the tribe Triticeae (Gramineae). Systematic Biology. 1996;45: 524–545. [Google Scholar]
- 37. Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 38. Simpson DW, Winterbottom CQ, Bell JA, Maltoni ML. Resistance to a single UK isolate of Colletotrichum acutatum in strawberry germplasm from Northern Europe. Euphytica. 1994;77: 161–164. [Google Scholar]
- 39. Denoyes-Rothan B, Lafargue M, Guerin G, Clerjeau M. Fruit resistance to Colletotrichum acutatum in strawberries. Plant Disease. 1999;83: 549–553. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
a 0, no visible lesions; 1, lesions on less than 33% of fruit surface; 2, lesions covering 33–66% of fruit surface; and 3, lesions covering more than 66% of fruit surface. b no lesions; 1, crown tissues discoloration but no wilting or collapse; 2, wilting or collapse of part of the plant; and 3, plant death.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.