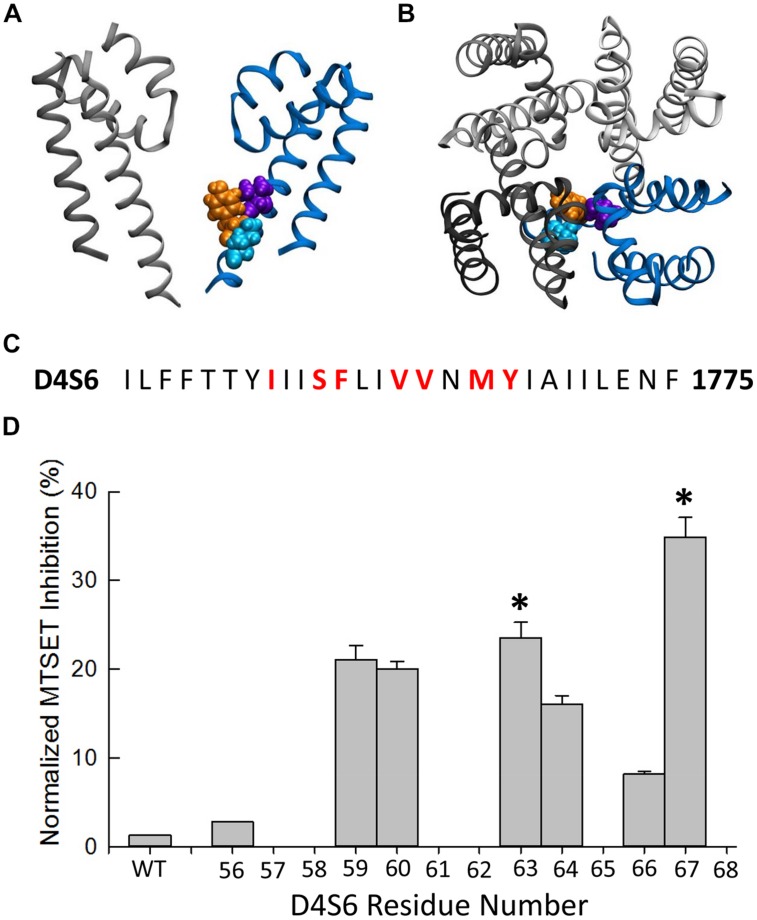
FIGURE 2.
Methanethiosulfonate reagent inhibition of D4S6 cysteine mutants. (A,B) Homology model of the Nav1.5 pore-forming region (See Materials and Methods). Model shows the orientation of the Nav1.5 D4S6 segment (blue) residues V1763 (purple), Y1767 (orange), and I1770 (cyan) viewed from the side (A) and top (B) of the channel. The D2 segment is shown in gray and the D1 (front) and D3 (back) segments were removed for clarity. (C) Primary amino acid sequence of the Nav1.5 D4S6 segment. Residues mutated to cysteine are shown in red. (D) The Na+ currents of D4S6 cysteine mutants were measured immediately after break in (P1) and after 5 min of internal 1 mM MTSET (P60). MTSET inhibition was calculated from the ratio of the P60 and P1 currents [1–(IP60/IP1)∗100] and plotted versus the D4S6 residue number (I1756–Y1767). Asterisks indicate significant differences in the MTSET inhibition of the Na+ currents of adjacent pairs of D4S6 cysteine mutants.